TriplEP-CPP: Algorithm for Predicting the Properties of Peptide Sequences
- PMID: 38999985
- PMCID: PMC11241344
- DOI: 10.3390/ijms25136869
TriplEP-CPP: Algorithm for Predicting the Properties of Peptide Sequences
Abstract
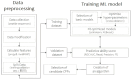
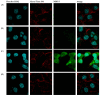
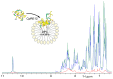
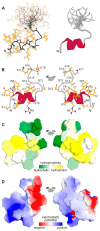
Advancements in medicine and pharmacology have led to the development of systems that deliver biologically active molecules inside cells, increasing drug concentrations at target sites. This improves effectiveness and duration of action and reduces side effects on healthy tissues. Cell-penetrating peptides (CPPs) show promise in this area. While traditional medicinal chemistry methods have been used to develop CPPs, machine learning techniques can speed up and reduce costs in the search for new peptides. A predictive algorithm based on machine learning models was created to identify novel CPP sequences using molecular descriptors using a combination of algorithms like k-nearest neighbors, gradient boosting, and random forest. Some potential CPPs were found and tested for cytotoxicity and penetrating ability. A new low-toxicity CPP was discovered from the Rhopilema esculentum venom proteome through this study.
Keywords: cell penetrating peptides (CPP); functional activity prediction; intracellular delivery; machine learning; protein-lipid interaction; structural-dynamic properties.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
KELM-CPPpred: Kernel Extreme Learning Machine Based Prediction Model for Cell-Penetrating Peptides.J Proteome Res. 2018 Sep 7;17(9):3214-3222. doi: 10.1021/acs.jproteome.8b00322. Epub 2018 Aug 13. J Proteome Res. 2018. PMID: 30032609
-
TargetCPP: accurate prediction of cell-penetrating peptides from optimized multi-scale features using gradient boost decision tree.J Comput Aided Mol Des. 2020 Aug;34(8):841-856. doi: 10.1007/s10822-020-00307-z. Epub 2020 Mar 16. J Comput Aided Mol Des. 2020. PMID: 32180124
-
Predicting cell-penetrating peptides using machine learning algorithms and navigating in their chemical space.Sci Rep. 2021 Apr 7;11(1):7628. doi: 10.1038/s41598-021-87134-w. Sci Rep. 2021. PMID: 33828175 Free PMC article.
-
The Development of Machine Learning Methods in Cell-Penetrating Peptides Identification: A Brief Review.Curr Drug Metab. 2019;20(3):217-223. doi: 10.2174/1389200219666181010114750. Curr Drug Metab. 2019. PMID: 30317992 Review.
-
Empirical comparison and analysis of web-based cell-penetrating peptide prediction tools.Brief Bioinform. 2020 Mar 23;21(2):408-420. doi: 10.1093/bib/bby124. Brief Bioinform. 2020. PMID: 30649170 Review.
Cited by
-
RoseTTAFold diffusion-guided short peptide design: a case study of binders against Keap1/Nrf2.Comput Struct Biotechnol J. 2025 Mar 1;27:896-911. doi: 10.1016/j.csbj.2025.02.032. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40123800 Free PMC article.
-
Exploring the Chemical Features and Biomedical Relevance of Cell-Penetrating Peptides.Int J Mol Sci. 2024 Dec 25;26(1):59. doi: 10.3390/ijms26010059. Int J Mol Sci. 2024. PMID: 39795918 Free PMC article. Review.
References
-
- Lindgren M., Langel Ü. Classes and Prediction of Cell-Penetrating Peptides. In: Langel Ü., editor. Cell-Penetrating Peptides. Volume 683. Humana Press; Totowa, NJ, USA: 2011. pp. 3–19. Methods in Molecular Biology. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

