TriplEP-CPP: Algorithm for Predicting the Properties of Peptide Sequences
- PMID: 38999985
- PMCID: PMC11241344
- DOI: 10.3390/ijms25136869
TriplEP-CPP: Algorithm for Predicting the Properties of Peptide Sequences
Abstract
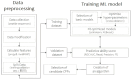
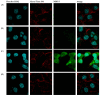
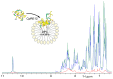
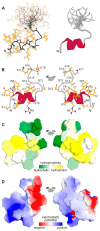
Advancements in medicine and pharmacology have led to the development of systems that deliver biologically active molecules inside cells, increasing drug concentrations at target sites. This improves effectiveness and duration of action and reduces side effects on healthy tissues. Cell-penetrating peptides (CPPs) show promise in this area. While traditional medicinal chemistry methods have been used to develop CPPs, machine learning techniques can speed up and reduce costs in the search for new peptides. A predictive algorithm based on machine learning models was created to identify novel CPP sequences using molecular descriptors using a combination of algorithms like k-nearest neighbors, gradient boosting, and random forest. Some potential CPPs were found and tested for cytotoxicity and penetrating ability. A new low-toxicity CPP was discovered from the Rhopilema esculentum venom proteome through this study.
Keywords: cell penetrating peptides (CPP); functional activity prediction; intracellular delivery; machine learning; protein-lipid interaction; structural-dynamic properties.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Lindgren M., Langel Ü. Classes and Prediction of Cell-Penetrating Peptides. In: Langel Ü., editor. Cell-Penetrating Peptides. Volume 683. Humana Press; Totowa, NJ, USA: 2011. pp. 3–19. Methods in Molecular Biology. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

