Deciphering the Interplay between the Epithelial Barrier, Immune Cells, and Metabolic Mediators in Allergic Disease
- PMID: 39000023
- PMCID: PMC11241838
- DOI: 10.3390/ijms25136913
Deciphering the Interplay between the Epithelial Barrier, Immune Cells, and Metabolic Mediators in Allergic Disease
Abstract
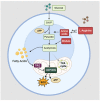
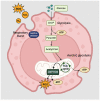
Chronic exposure to harmful pollutants, chemicals, and pathogens from the environment can lead to pathological changes in the epithelial barrier, which increase the risk of developing an allergy. During allergic inflammation, epithelial cells send proinflammatory signals to group 2 innate lymphoid cell (ILC2s) and eosinophils, which require energy and resources to mediate their activation, cytokine/chemokine secretion, and mobilization of other cells. This review aims to provide an overview of the metabolic regulation in allergic asthma, atopic dermatitis (AD), and allergic rhinitis (AR), highlighting its underlying mechanisms and phenotypes, and the potential metabolic regulatory roles of eosinophils and ILC2s. Eosinophils and ILC2s regulate allergic inflammation through lipid mediators, particularly cysteinyl leukotrienes (CysLTs) and prostaglandins (PGs). Arachidonic acid (AA)-derived metabolites and Sphinosine-1-phosphate (S1P) are significant metabolic markers that indicate immune dysfunction and epithelial barrier dysfunction in allergy. Notably, eosinophils are promoters of allergic symptoms and exhibit greater metabolic plasticity compared to ILC2s, directly involved in promoting allergic symptoms. Our findings suggest that metabolomic analysis provides insights into the complex interactions between immune cells, epithelial cells, and environmental factors. Potential therapeutic targets have been highlighted to further understand the metabolic regulation of eosinophils and ILC2s in allergy. Future research in metabolomics can facilitate the development of novel diagnostics and therapeutics for future application.
Keywords: ILC2; allergic inflammation; allergy; eosinophils; lipid mediators; metabolites.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

