Uncovering PheCLE1 and PheCLE10 Promoting Root Development Based on Genome-Wide Analysis
- PMID: 39000298
- PMCID: PMC11241622
- DOI: 10.3390/ijms25137190
Uncovering PheCLE1 and PheCLE10 Promoting Root Development Based on Genome-Wide Analysis
Abstract
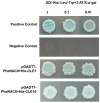
Moso bamboo (Phyllostachys edulis), renowned for its rapid growth, is attributed to the dynamic changes in its apical meristem. The CLAVATA3/EMBRYO SURROUNDING REGION-RELATED (CLE) family genes are known to play crucial roles in regulating meristem and organ formation in model plants, but their functions in Moso bamboo remain unclear. Here, we conducted a genome-wide identification of the CLE gene family of Moso bamboo and investigated their gene structure, chromosomal localization, evolutionary relationships, and expression patterns. A total of 11 PheCLE genes were identified, all of which contained a conserved CLE peptide core functional motif (Motif 1) at their C-termini. Based on Arabidopsis classification criteria, these genes were predominantly distributed in Groups A-C. Collinearity analysis unveiled significant synteny among CLE genes in Moso bamboo, rice, and maize, implying potential functional conservation during monocot evolution. Transcriptomic analysis showed significant expression of these genes in the apical tissues of Moso bamboo, including root tips, shoot tips, rhizome buds, and flower buds. Particularly, single-cell transcriptomic data and in situ hybridization further corroborated the heightened expression of PheCLE1 and PheCLE10 in the apical tissue of basal roots. Additionally, the overexpression of PheCLE1 and PheCLE10 in rice markedly promoted root growth. PheCLE1 and PheCLE10 were both located on the cell membrane. Furthermore, the upstream transcription factors NAC9 and NAC6 exhibited binding affinity toward the promoters of PheCLE1 and PheCLE10, thereby facilitating their transcriptional activation. In summary, this study not only systematically identified the CLE gene family in Moso bamboo for the first time but also emphasized their central roles in apical tissue development. This provides a valuable theoretical foundation for the further exploration of functional peptides and their signaling regulatory networks in bamboo species.
Keywords: CLE gene family; Moso bamboo; PheCLE1; PheCLE10; apical tissue development.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







References
-
- INBAR . 2022 Global Bamboo and Rattan Commodities International Trade Report. International Bamboo and Rattan Organization; Beijing, China: 2023. [(accessed on 25 April 2024)]. Available online: https://www.inbar.int/resources/inbar_publications/2022zgztspgjmybg/
-
- Yang J.B., Dong Y.R., Wong K.M., Gu Z.J., Yang H.Q., Li D.Z. Genetic structure and differentiation in Dendrocalamus sinicus (Poaceae: Bambusoideae) populations provide insight into evolutionary history and speciation of woody bamboos. Sci. Rep. 2018;8:16933. doi: 10.1038/s41598-018-35269-8. - DOI - PMC - PubMed
-
- Iroegbu A.O.C., Ray S.S. Bamboos: From bioresource to sustainable materials and chemicals. Sustainability. 2021;13:12200. doi: 10.3390/su132112200. - DOI
-
- Zhou G., Gu L., Chai Q., Zhang J., Zhu W., Zhang Z. Carbon footprints labeling of bamboo products: A powerful tool to promote innovative development of bamboo industry. World Bamboo Ratt. 2022;20:29.
-
- Chen M., Guo L., Ramakrishnan M., Fei Z., Vinod K.K., Ding Y., Jiao C., Gao Z., Zha R., Wang C., et al. Rapid growth of Moso bamboo (Phyllostachys edulis): Cellular roadmaps, transcriptome dynamics, and environmental factors. Plant Cell. 2022;34:3577–3610. doi: 10.1093/plcell/koac193. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

