ADAR-Mediated A>I(G) RNA Editing in the Genotoxic Drug Response of Breast Cancer
- PMID: 39000531
- PMCID: PMC11242177
- DOI: 10.3390/ijms25137424
ADAR-Mediated A>I(G) RNA Editing in the Genotoxic Drug Response of Breast Cancer
Abstract
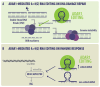
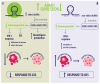
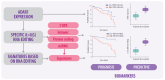
Epitranscriptomics is a field that delves into post-transcriptional changes. Among these modifications, the conversion of adenosine to inosine, traduced as guanosine (A>I(G)), is one of the known RNA-editing mechanisms, catalyzed by ADARs. This type of RNA editing is the most common type of editing in mammals and contributes to biological diversity. Disruption in the A>I(G) RNA-editing balance has been linked to diseases, including several types of cancer. Drug resistance in patients with cancer represents a significant public health concern, contributing to increased mortality rates resulting from therapy non-responsiveness and disease progression, representing the greatest challenge for researchers in this field. The A>I(G) RNA editing is involved in several mechanisms over the immunotherapy and genotoxic drug response and drug resistance. This review investigates the relationship between ADAR1 and specific A>I(G) RNA-edited sites, focusing particularly on breast cancer, and the impact of these sites on DNA damage repair and the immune response over anti-cancer therapy. We address the underlying mechanisms, bioinformatics, and in vitro strategies for the identification and validation of A>I(G) RNA-edited sites. We gathered databases related to A>I(G) RNA editing and cancer and discussed the potential clinical and research implications of understanding A>I(G) RNA-editing patterns. Understanding the intricate role of ADAR1-mediated A>I(G) RNA editing in breast cancer holds significant promise for the development of personalized treatment approaches tailored to individual patients' A>I(G) RNA-editing profiles.
Keywords: A>I(G); ADAR1; DNA damage repair; RNA editing; breast cancer; drug resistance; drug response; immune response; splicing alteration.
Conflict of interest statement
R.A. declares honoraria for conferences, advisory boards, and educational activities from Roche, grants, and support for scientific research from Illumina, Pfizer, Roche, and Thermo Fisher Scientific, and honoraria for conferences from Thermo Fisher Scientific, Janssen, and Tecnofarma. The other authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

