Enhancing Medical Imaging Segmentation with GB-SAM: A Novel Approach to Tissue Segmentation Using Granular Box Prompts
- PMID: 39001452
- PMCID: PMC11240495
- DOI: 10.3390/cancers16132391
Enhancing Medical Imaging Segmentation with GB-SAM: A Novel Approach to Tissue Segmentation Using Granular Box Prompts
Abstract
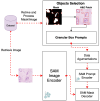
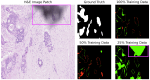
Recent advances in foundation models have revolutionized model development in digital pathology, reducing dependence on extensive manual annotations required by traditional methods. The ability of foundation models to generalize well with few-shot learning addresses critical barriers in adapting models to diverse medical imaging tasks. This work presents the Granular Box Prompt Segment Anything Model (GB-SAM), an improved version of the Segment Anything Model (SAM) fine-tuned using granular box prompts with limited training data. The GB-SAM aims to reduce the dependency on expert pathologist annotators by enhancing the efficiency of the automated annotation process. Granular box prompts are small box regions derived from ground truth masks, conceived to replace the conventional approach of using a single large box covering the entire H&E-stained image patch. This method allows a localized and detailed analysis of gland morphology, enhancing the segmentation accuracy of individual glands and reducing the ambiguity that larger boxes might introduce in morphologically complex regions. We compared the performance of our GB-SAM model against U-Net trained on different sizes of the CRAG dataset. We evaluated the models across histopathological datasets, including CRAG, GlaS, and Camelyon16. GB-SAM consistently outperformed U-Net, with reduced training data, showing less segmentation performance degradation. Specifically, on the CRAG dataset, GB-SAM achieved a Dice coefficient of 0.885 compared to U-Net's 0.857 when trained on 25% of the data. Additionally, GB-SAM demonstrated segmentation stability on the CRAG testing dataset and superior generalization across unseen datasets, including challenging lymph node segmentation in Camelyon16, which achieved a Dice coefficient of 0.740 versus U-Net's 0.491. Furthermore, compared to SAM-Path and Med-SAM, GB-SAM showed competitive performance. GB-SAM achieved a Dice score of 0.900 on the CRAG dataset, while SAM-Path achieved 0.884. On the GlaS dataset, Med-SAM reported a Dice score of 0.956, whereas GB-SAM achieved 0.885 with significantly less training data. These results highlight GB-SAM's advanced segmentation capabilities and reduced dependency on large datasets, indicating its potential for practical deployment in digital pathology, particularly in settings with limited annotated datasets.
Keywords: digital pathology; foundation models; histopathology; pathology image; segmentation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures









References
-
- Farahani N., Parwani A.V., Pantanowitz L. Whole slide imaging in pathology: Advantages, limitations, and emerging perspectives. Pathol. Lab. Med. Int. 2015;2015:23–33.
-
- Huang Z., Yang E., Shen J., Gratzinger D., Eyerer F., Liang B., Nirschl J., Bingham D., Dussaq A.M., Kunder C., et al. A pathologist–AI collaboration framework for enhancing diagnostic accuracies and efficiencies. Nat. Biomed. Eng. 2024:1–16. doi: 10.1038/s41551-024-01223-5. Online ahead of print . - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

