Characterisation of LPS+ bacterial extracellular vesicles along the gut-hepatic portal vein-liver axis
- PMID: 39001704
- PMCID: PMC11245684
- DOI: 10.1002/jev2.12474
Characterisation of LPS+ bacterial extracellular vesicles along the gut-hepatic portal vein-liver axis
Abstract
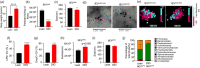
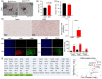
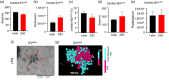
Gut microbiome dysbiosis is a major contributing factor to several pathological conditions. However, the mechanistic understanding of the communication between gut microbiota and extra-intestinal organs remains largely elusive. Extracellular vesicles (EVs), secreted by almost every form of life, including bacteria, could play a critical role in this inter-kingdom crosstalk and are the focus of present study. Here, we present a novel approach for isolating lipopolysaccharide (LPS)+ bacterial extracellular vesicles (bEVLPS) from complex biological samples, including faeces, plasma and the liver from lean and diet-induced obese (DIO) mice. bEVLPS were extensively characterised using nanoparticle tracking analyses, immunogold labelling coupled with transmission electron microscopy, flow cytometry, super-resolution microscopy and 16S sequencing. In liver tissues, the protein expressions of TLR4 and a few macrophage-specific biomarkers were assessed by immunohistochemistry, and the gene expressions of inflammation-related cytokines and their receptors (n = 89 genes) were measured using a PCR array. Faecal samples from DIO mice revealed a remarkably lower concentration of total EVs but a significantly higher percentage of LPS+ EVs. Interestingly, DIO faecal bEVLPS showed a higher abundance of Proteobacteria by 16S sequencing. Importantly, in DIO mice, a higher number of total EVs and bEVLPS consistently entered the hepatic portal vein and subsequently reached the liver, associated with increased expression of TLR4, macrophage markers (F4/80, CD86 and CD206), cytokines and receptors (Il1rn, Ccr1, Cxcl10, Il2rg and Ccr2). Furthermore, a portion of bEVLPS escaped liver and entered the peripheral circulation. In conclusion, bEV could be the key mediator orchestrating various well-established biological effects induced by gut bacteria on distant organs.
Keywords: bacterial extracellular vesicles; dysbiosis; gut microbiome; inflammation; lipopolysaccharide.
© 2024 The Author(s). Journal of Extracellular Vesicles published by Wiley Periodicals LLC on behalf of International Society for Extracellular Vesicles.
Conflict of interest statement
GD is the founder of LiBiCo that has no influence or contribution to the work presented in this manuscript.
Figures


References
-
- Avila‐Calderón, E. D. , Ruiz‐Palma, M. D. S. , Aguilera‐Arreola, M. G. , Velázquez‐Guadarrama, N. , Ruiz, E. A. , Gomez‐Lunar, Z. , Witonsky, S. , & Contreras‐Rodríguez, A. (2021). Outer membrane vesicles of gram‐negative bacteria: An outlook on biogenesis. Frontiers in Microbiology, 12, 557902. 10.3389/fmicb.2021.557902 - DOI - PMC - PubMed
-
- Bartneck, M. , Koppe, C. , Fech, V. , Warzecha, K. T. , Kohlhepp, M. , Huss, S. , Weiskirchen, R. , Trautwein, C. , Luedde, T. , & Tacke, F. (2021). Roles of CCR2 and CCR5 for hepatic macrophage polarization in mice with liver parenchymal cell‐specific NEMO deletion. Cellular and Molecular Gastroenterology and Hepatology, 11(2), 327–347. 10.1016/j.jcmgh.2020.08.012 - DOI - PMC - PubMed
-
- Batut, B. , Hiltemann, S. , Bagnacani, A. , Baker, D. , Bhardwaj, V. , Blank, C. , Bretaudeau, A. , Brillet‐Guéguen, L. , Čech, M. , Chilton, J. , Clements, D. , Doppelt‐Azeroual, O. , Erxleben, A. , Freeberg, M. A. , Gladman, S. , Hoogstrate, Y. , Hotz, H. R. , Houwaart, T. , Jagtap, P. , … Grüning, B. (2018). Community‐driven data analysis training for biology. Cell Systems, 6(6), 752–758.e1. 10.1016/j.cels.2018.05.012 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

