Early human fetal lung atlas reveals the temporal dynamics of epithelial cell plasticity
- PMID: 39003323
- PMCID: PMC11246468
- DOI: 10.1038/s41467-024-50281-5
Early human fetal lung atlas reveals the temporal dynamics of epithelial cell plasticity
Abstract
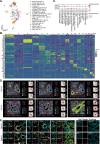
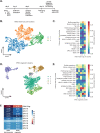
Studying human fetal lungs can inform how developmental defects and disease states alter the function of the lungs. Here, we sequenced >150,000 single cells from 19 healthy human pseudoglandular fetal lung tissues ranging between gestational weeks 10-19. We capture dynamic developmental trajectories from progenitor cells that express abundant levels of the cystic fibrosis conductance transmembrane regulator (CFTR). These cells give rise to multiple specialized epithelial cell types. Combined with spatial transcriptomics, we show temporal regulation of key signalling pathways that may drive the temporal and spatial emergence of specialized epithelial cells including ciliated and pulmonary neuroendocrine cells. Finally, we show that human pluripotent stem cell-derived fetal lung models contain CFTR-expressing progenitor cells that capture similar lineage developmental trajectories as identified in the native tissue. Overall, this study provides a comprehensive single-cell atlas of the developing human lung, outlining the temporal and spatial complexities of cell lineage development and benchmarks fetal lung cultures from human pluripotent stem cell differentiations to similar developmental window.
© 2024. The Author(s).
Conflict of interest statement
All authors declare no competing interests.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

