A bipolar disorder-associated missense variant alters adenylyl cyclase 2 activity and promotes mania-like behavior
- PMID: 39003412
- PMCID: PMC11649569
- DOI: 10.1038/s41380-024-02663-w
A bipolar disorder-associated missense variant alters adenylyl cyclase 2 activity and promotes mania-like behavior
Abstract
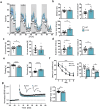
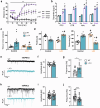
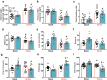
The single nucleotide polymorphism rs13166360, causing a substitution of valine (Val) 147 to leucine (Leu) in the adenylyl cyclase 2 (ADCY2), has previously been associated with bipolar disorder (BD). Here we show that the disease-associated ADCY2 missense mutation diminishes the enzyme´s capacity to generate the second messenger 3',5'-cylic adenosine monophosphate (cAMP) by altering its subcellular localization. We established mice specifically carrying the Val to Leu substitution using CRISPR/Cas9-based gene editing. Mice homozygous for the Leu variant display symptoms of a mania-like state accompanied by cognitive impairments. Mutant animals show additional characteristic signs of rodent mania models, i.e., they are hypersensitive to amphetamine, the observed mania-like behaviors are responsive to lithium treatment and the Val to Leu substitution results in a shifted excitatory/inhibitory synaptic balance towards more excitation. Exposure to chronic social defeat stress switches homozygous Leu variant carriers from a mania- to a depressive-like state, a transition which is reminiscent of the alternations characterizing the symptomatology in BD patients. Single-cell RNA-seq (scRNA-seq) revealed widespread Adcy2 mRNA expression in numerous hippocampal cell types. Differentially expressed genes particularly identified from glutamatergic CA1 neurons point towards ADCY2 variant-dependent alterations in multiple biological processes including cAMP-related signaling pathways. These results validate ADCY2 as a BD risk gene, provide insights into underlying disease mechanisms, and potentially open novel avenues for therapeutic intervention strategies.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval: All animal experiments were conducted with the approval of and in accordance with the Guide of the Care and Use of Laboratory Animals of the Government of Upper Bavaria, Germany.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

