DNA Methylation-derived biological age and long-term mortality risk in subjects with type 2 diabetes
- PMID: 39003492
- PMCID: PMC11245869
- DOI: 10.1186/s12933-024-02351-7
DNA Methylation-derived biological age and long-term mortality risk in subjects with type 2 diabetes
Abstract
Background: Individuals with type 2 diabetes (T2D) face an increased mortality risk, not fully captured by canonical risk factors. Biological age estimation through DNA methylation (DNAm), i.e. the epigenetic clocks, is emerging as a possible tool to improve risk stratification for multiple outcomes. However, whether these tools predict mortality independently of canonical risk factors in subjects with T2D is unknown.
Methods: Among a cohort of 568 T2D patients followed for 16.8 years, we selected a subgroup of 50 subjects, 27 survived and 23 deceased at present, passing the quality check and balanced for all risk factors after propensity score matching. We analyzed DNAm from peripheral blood leukocytes using the Infinium Human MethylationEPIC BeadChip (Illumina) to evaluate biological aging through previously validated epigenetic clocks and assess the DNAm-estimated levels of selected inflammatory proteins and blood cell counts. We tested the associations of these estimates with mortality using two-stage residual-outcome regression analysis, creating a reference model on data from the group of survived patients.
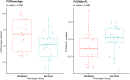
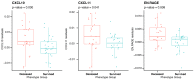
Results: Deceased subjects had higher median epigenetic age expressed with DNAmPhenoAge algorithm (57.49 [54.72; 60.58] years. vs. 53.40 [49.73; 56.75] years; p = 0.012), and accelerated DunedinPoAm pace of aging (1.05 [1.02; 1.11] vs. 1.02 [0.98; 1.06]; p = 0.012). DNAm PhenoAge (HR 1.16, 95% CI 1.05-1.28; p = 0.004) and DunedinPoAm (HR 3.65, 95% CI 1.43-9.35; p = 0.007) showed an association with mortality independently of canonical risk factors. The epigenetic predictors of 3 chronic inflammation-related proteins, i.e. CXCL10, CXCL11 and enRAGE, C-reactive protein methylation risk score and DNAm-based estimates of exhausted CD8 + T cell counts were higher in deceased subjects when compared to survived.
Conclusions: These findings suggest that biological aging, as estimated through existing epigenetic tools, is associated with mortality risk in individuals with T2D, independently of common risk factors and that increased DNAm-surrogates of inflammatory protein levels characterize deceased T2D patients. Replication in larger cohorts is needed to assess the potential of this approach to refine mortality risk in T2D.
Keywords: DNA methylation; DunedinPoAm; Epigenetic clocks; PhenoAge; Type 2 diabetes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Wright AK, Suarez-Ortegon MF, Read SH, Kontopantelis E, Buchan I, Emsley R, et al. Risk factor control and cardiovascular event risk in people with type 2 diabetes in primary and secondary prevention settings. Circulation. 2020;142(20):1925–1936. doi: 10.1161/CIRCULATIONAHA.120.046783. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

