Investigation of cannabidiol-induced cytotoxicity in human hepatic cells
- PMID: 39004336
- PMCID: PMC11648445
- DOI: 10.1016/j.tox.2024.153884
Investigation of cannabidiol-induced cytotoxicity in human hepatic cells
Abstract
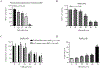
Cannabidiol (CBD) is one of the primary cannabinoids present in extracts of the plant Cannabis sativa L. A CBD-based drug, Epidiolex, has been approved by the U.S. FDA for the treatment of seizures in childhood-onset epileptic disorders. Although CBD-associated liver toxicity has been reported in clinical studies, the underlying mechanisms remain unclear. In this study, we demonstrated that CBD causes cytotoxicity in primary human hepatocytes and hepatic HepG2 cells. A 24-h CBD treatment induced cell cycle disturbances, cellular apoptosis, and endoplasmic reticulum (ER) stress in HepG2 cells. A potent ER stress inhibitor, 4-phenylbutyrate, markedly attenuated CBD-induced apoptosis and cell death. Additionally, we investigated the role of cytochrome P450 (CYP)-mediated metabolism in CBD-induced cytotoxicity using HepG2 cell lines engineered to express 14 individual CYPs. We identified CYP2C9, 2C19, 2D6, 2C18, and 3A5 as participants in CBD metabolism. Notably, cells overexpressing CYP2C9, 2C19, and 2C18 produced 7-hydroxy-CBD, while cells overexpressing CYP2C9, 2C19, 2D6, and 2C18 generated 7-carboxy-CBD. Furthermore, CBD-induced cytotoxicity was significantly attenuated in the cells expressing CYP2D6. Taken together, these data suggest that cell cycle disturbances, apoptosis, and ER stress are associated with CBD-induced cytotoxicity, and CYP2D6-mediated metabolism plays a critical role in decreasing the cytotoxicity of CBD.
Keywords: Apoptosis; CBD; CYPs; ER stress; Liver toxicity; Topoisomerases.
Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
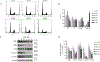



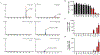

References
-
- Aquilina G, Crescenzi M and Bignami M 1999. Mismatch repair, G(2)/M cell cycle arrest and lethality after DNA damage. Carcinogenesis 20, 2317–2326. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

