YAP/TAZ interacts with RBM39 to confer resistance against indisulam
- PMID: 39004623
- PMCID: PMC11247092
- DOI: 10.1038/s41389-024-00527-0
YAP/TAZ interacts with RBM39 to confer resistance against indisulam
Abstract
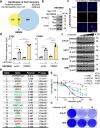
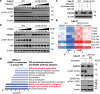
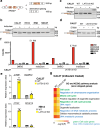
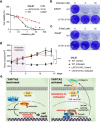
The Hippo pathway and its downstream effectors, Yes-associated protein/transcriptional coactivator with PDZ-binding motif (YAP/TAZ), are essential for cell growth and organ development. Emerging evidence revealed that the Hippo pathway and YAP/TAZ are frequently dysregulated by multiple genetic alterations in solid cancers including head and neck squamous cell carcinoma (HNSCC); however, the YAP/TAZ-nuclear interactome remains unclear. RNA-binding motif protein 39 (RBM39) enhances transcriptional activity of several transcription factors and also regulates mRNA splicing. Indisulam degrading RBM39 induces alternative splicing, leading to cell death. However, clinical trials of indisulam have failed to show effectiveness. Therefore, clarifying the resistance mechanism against splicing inhibitors is urgently required. In this study, we identified RBM39 as a novel YAP/TAZ-interacting molecule by proteome analysis. RBM39 promoted YAP/TAZ transcriptional activity. We further elucidated that indisulam reduces RBM39/YAP/TAZ-mediated integrin or collagen expression, thereby inactivating focal adhesion kinase (FAK) important for cell survival. Moreover, indisulam also induced alternative splicing of cell cycle- or DNA metabolism-related genes. YAP/TAZ hyperactivation delayed indisulam-induced RBM39 degradation, which restored the integrin/collagen expression to activate FAK, and alternative splicing, thereby conferring resistance against indisulam in vitro and in vivo. Our findings may aid to develop a novel cancer therapy focusing on YAP/TAZ/RBM39 interaction.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

