Novel Long Noncoding RNA HEAT4 Affects Monocyte Subtypes, Reducing Inflammation and Promoting Vascular Healing
- PMID: 39005211
- PMCID: PMC11444369
- DOI: 10.1161/CIRCULATIONAHA.124.069315
Novel Long Noncoding RNA HEAT4 Affects Monocyte Subtypes, Reducing Inflammation and Promoting Vascular Healing
Abstract
Background: Activation of the immune system contributes to cardiovascular diseases. The role of human-specific long noncoding RNAs in cardioimmunology is poorly understood.
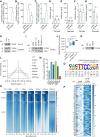
Methods: Single-cell sequencing in peripheral blood mononuclear cells revealed a novel human-specific long noncoding RNA called HEAT4 (heart failure-associated transcript 4). HEAT4 expression was assessed in several in vitro and ex vivo models of immune cell activation, as well as in the blood of patients with heart failure (HF), acute myocardial infarction, or cardiogenic shock. The transcriptional regulation of HEAT4 was verified through cytokine treatment and single-cell sequencing. Loss-of-function and gain-of-function studies and multiple RNA-protein interaction assays uncovered a mechanistic role of HEAT4 in the monocyte anti-inflammatory gene program. HEAT4 expression and function was characterized in a vascular injury model in NOD.CB17-Prkdc scid/Rj mice.
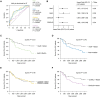
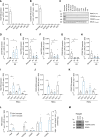
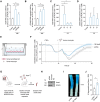
Results: HEAT4 expression was increased in the blood of patients with HF, acute myocardial infarction, or cardiogenic shock. HEAT4 levels distinguished patients with HF from people without HF and predicted all-cause mortality in a cohort of patients with HF over 7 years of follow-up. Monocytes, particularly anti-inflammatory CD16+ monocytes, which are increased in patients with HF, are the primary source of HEAT4 expression in the blood. HEAT4 is transcriptionally activated by treatment with anti-inflammatory interleukin-10. HEAT4 activates anti-inflammatory and inhibits proinflammatory gene expression. Increased HEAT4 levels result in a shift toward more CD16+ monocytes. HEAT4 binds to S100A9, causing a monocyte subtype switch, thereby reducing inflammation. As a result, HEAT4 improves endothelial barrier integrity during inflammation and promotes vascular healing after injury in mice.
Conclusions: These results characterize a novel endogenous anti-inflammatory pathway that involves the conversion of monocyte subtypes into anti-inflammatory CD16+ monocytes. The data identify a novel function for the class of long noncoding RNAs by preventing protein secretion and suggest long noncoding RNAs as potential targets for interventions in the field of cardioimmunology.
Keywords: RNA, long noncoding; heart failure; monocytes; regeneration.
Conflict of interest statement
None.
Figures




References
-
- Greten FR, Eckmann L, Greten TF, Park JM, Li Z-W, Egan LJ, Kagnoff MF, Karin M. IKKβ links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013 - PubMed
-
- Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, Gutkovich-Pyest E, Urieli-Shoval S, Galun E, Ben-Neriah Y. NF-κB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–466. doi: 10.1038/nature02924 - PubMed
-
- McInnes IB, Schett G. Cytokines in the pathogenesis of rheumatoid arthritis. Nat Rev Immunol. 2007;7:429–442. doi: 10.1038/nri2094 - PubMed
-
- Levine B, Kalman J, Mayer L, Fillit HM, Packer M. Elevated circulating levels of tumor necrosis factor in severe chronic heart failure. N Engl J Med. 1990;323:236–241. doi: 10.1056/NEJM199007263230405 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

