This is a preprint.
The Neurolipid Atlas: a lipidomics resource for neurodegenerative diseases uncovers cholesterol as a regulator of astrocyte reactivity impaired by ApoE4
- PMID: 39005258
- PMCID: PMC11244892
- DOI: 10.1101/2024.07.01.601474
The Neurolipid Atlas: a lipidomics resource for neurodegenerative diseases uncovers cholesterol as a regulator of astrocyte reactivity impaired by ApoE4
Update in
-
The Neurolipid Atlas: a lipidomics resource for neurodegenerative diseases.Nat Metab. 2025 Oct;7(10):2142-2164. doi: 10.1038/s42255-025-01365-z. Epub 2025 Sep 22. Nat Metab. 2025. PMID: 40983680 Free PMC article.
Abstract
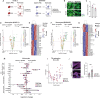
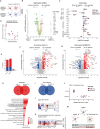
Lipid changes in the brain have been implicated in many neurodegenerative diseases including Alzheimer's Disease (AD), Parkinson's disease and Amyotrophic Lateral Sclerosis. To facilitate comparative lipidomic research across brain-diseases we established a data commons named the Neurolipid Atlas, that we have pre-populated with novel human, mouse and isogenic induced pluripotent stem cell (iPSC)-derived lipidomics data for different brain diseases. We show that iPSC-derived neurons, microglia and astrocytes display distinct lipid profiles that recapitulate in vivo lipotypes. Leveraging multiple datasets, we show that the AD risk gene ApoE4 drives cholesterol ester (CE) accumulation in human astrocytes recapitulating CE accumulation measured in the human AD brain. Multi-omic interrogation of iPSC-derived astrocytes revealed that cholesterol plays a major role in astrocyte interferon-dependent pathways such as the immunoproteasome and major histocompatibility complex (MHC) class I antigen presentation. We show that through enhanced cholesterol esterification ApoE4 suppresses immune activation of astrocytes. Our novel data commons, available at neurolipidatlas.com, provides a user-friendly tool and knowledge base for a better understanding of lipid dyshomeostasis in neurodegenerative diseases.
Figures





References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
