This is a preprint.
Functional analysis of ESRP1/2 gene variants and CTNND1 isoforms in orofacial cleft pathogenesis
- PMID: 39005284
- PMCID: PMC11245018
- DOI: 10.1101/2024.07.02.601574
Functional analysis of ESRP1/2 gene variants and CTNND1 isoforms in orofacial cleft pathogenesis
Update in
-
Functional analysis of ESRP1/2 gene variants and CTNND1 isoforms in orofacial cleft pathogenesis.Commun Biol. 2024 Aug 23;7(1):1040. doi: 10.1038/s42003-024-06715-3. Commun Biol. 2024. PMID: 39179789 Free PMC article.
Abstract
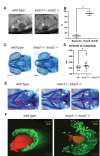
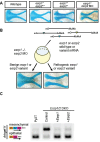
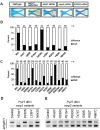
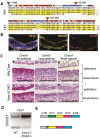
Orofacial cleft (OFC) is a common human congenital anomaly. Epithelial-specific RNA splicing regulators ESRP1 and ESRP2 regulate craniofacial morphogenesis and their disruption result in OFC in zebrafish, mouse and humans. Using esrp1/2 mutant zebrafish and murine Py2T cell line models, we functionally tested the pathogenicity of human ESRP1/2 gene variants. We found that many variants predicted by in silico methods to be pathogenic were functionally benign. Esrp1 also regulates the alternative splicing of Ctnnd1 and these genes are co-expressed in the embryonic and oral epithelium. In fact, over-expression of ctnnd1 is sufficient to rescue morphogenesis of epithelial-derived structures in esrp1/2 zebrafish mutants. Additionally, we identified 13 CTNND1 variants from genome sequencing of OFC cohorts, confirming CTNND1 as a key gene in human OFC. This work highlights the importance of functional assessment of human gene variants and demonstrates the critical requirement of Esrp-Ctnnd1 acting in the embryonic epithelium to regulate palatogenesis.
Figures


References
-
- Leslie EJ. Genetic models and approaches to study orofacial clefts. Oral Dis. 2021. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
