This is a preprint.
Influenza A defective viral genome production is altered by metabolites, metabolic signaling molecules, and cyanobacteria extracts
- PMID: 39005323
- PMCID: PMC11245085
- DOI: 10.1101/2024.07.04.602134
Influenza A defective viral genome production is altered by metabolites, metabolic signaling molecules, and cyanobacteria extracts
Abstract
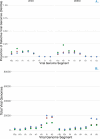
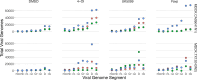
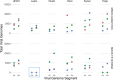
RNA virus infections are composed of a diverse mix of viral genomes that arise from low fidelity in replication within cells. The interactions between "defective" and full-length viral genomes have been shown to shape pathogenesis, leading to intense research into employing these to develop novel antivirals. In particular, Influenza A defective viral genomes (DVGs) have been associated with milder clinical outcomes. Yet, the full potential of DVGs as broad-spectrum antivirals remains untapped due to the unknown mechanisms of their de novo production. Much of the research into the factors affecting defective viral genome production has focused on the virus, while the role of the host has been neglected. We recently showed that altering host cell metabolism away from pro-growth pathways using alpelisib increased the production of Influenza A defective viral genomes. To uncover other drugs that could induce infections to create more DVGs, we subjected active influenza infections of the two circulating human subtypes (A/H1N1 & A/H3N2) to a screen of metabolites, metabolic signaling molecules, and cyanobacteria-derived biologics, after which we quantified the defective viral genomes (specifically deletion-containing viral genomes, DelVGs) and total viral genomes using third generation long-read sequencing. Here we show that metabolites and signaling molecules of host cell central carbon metabolism can significantly alter DelVG production early in Influenza A infection. Adenosine, emerged as a potent inducer of defective viral genomes, significantly amplifying DelVG production across both subtypes. Insulin had similar effects, albeit subtype-specific, predominantly enhancing polymerase segment DVGs in TX12 infections. Tricarboxylic Acid (TCA) cycle inhibitors 4-octyl itaconate and UK5099, along with the purine analog favipiravir, increased total viral genome production across subtypes. Cyanobacterial extracts primarily affected DVG and total viral genome production in TX12, with a specific, almost complete shutdown of influenza antigenic segments. These results underscore the influence of host metabolic pathways on DVG production and suggest new avenues for antiviral intervention, including PI3K-AKT and Ras-MAPK signaling pathways, TCA cycle metabolism, purine-pyrimidine metabolism, polymerase inhibition, and cyanotherapeutic approaches. More broadly, our findings suggest that the social interactions observed between defective and full-length viral genomes, depend not only on the viral actors, but can be altered by the stage provided by the host. Our study advances our fundamental understanding of DVG production mechanisms and highlights the potential of targeting host metabolism to develop broad-spectrum influenza therapeutics.
Figures


Similar articles
-
Influenza A defective viral genomes and non-infectious particles are increased by host PI3K inhibition via anti-cancer drug alpelisib.bioRxiv [Preprint]. 2024 Jul 3:2024.07.03.601932. doi: 10.1101/2024.07.03.601932. bioRxiv. 2024. PMID: 39005364 Free PMC article. Preprint.
-
Levels of Influenza A Virus Defective Viral Genomes Determine Pathogenesis in the BALB/c Mouse Model.J Virol. 2022 Nov 9;96(21):e0117822. doi: 10.1128/jvi.01178-22. Epub 2022 Oct 13. J Virol. 2022. PMID: 36226985 Free PMC article.
-
Defective Viral Genomes Alter How Sendai Virus Interacts with Cellular Trafficking Machinery, Leading to Heterogeneity in the Production of Viral Particles among Infected Cells.J Virol. 2019 Feb 5;93(4):e01579-18. doi: 10.1128/JVI.01579-18. Print 2019 Feb 15. J Virol. 2019. PMID: 30463965 Free PMC article.
-
Defective viral genomes: advances in understanding their generation, function, and impact on infection outcomes.mBio. 2024 May 8;15(5):e0069224. doi: 10.1128/mbio.00692-24. Epub 2024 Apr 3. mBio. 2024. PMID: 38567955 Free PMC article. Review.
-
The Impact of Defective Viruses on Infection and Immunity.Annu Rev Virol. 2019 Sep 29;6(1):547-566. doi: 10.1146/annurev-virology-092818-015652. Epub 2019 May 13. Annu Rev Virol. 2019. PMID: 31082310 Review.
References
-
- Ackermann WW, Klernschmidt E. Concerning the relation of the Krebs cycle to virus propagation. J Biol Chem. 1951. Mar;189(1):421–8. - PubMed
-
- Agu I, José IR, Ram A, Oberbauer D, Albeck J, Díaz Muñoz SL. Influenza A defective viral genomes and non-infectious particles are increased by host PI3K inhibition via anti-cancer drug alpelisib. bioRxiv. 2024. 2024.07.03.601932; doi: 10.1101/2024.07.03.601932 - DOI
-
- Alnaji FG, Holmes JR, Rendon G, Vera JC, Fields CJ, Martin BE, Brooke CB. Sequencing Framework for the Sensitive Detection and Precise Mapping of Defective Interfering Particle-Associated Deletions across Influenza A and B Viruses. J Virol. 2019. May 15;93(11):e00354–19. doi: 10.1128/JVI.00354-19. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
