This is a preprint.
Epigenetic changes regulating the epithelial-mesenchymal transition in human trophoblast differentiation
- PMID: 39005325
- PMCID: PMC11244995
- DOI: 10.1101/2024.07.02.601748
Epigenetic changes regulating the epithelial-mesenchymal transition in human trophoblast differentiation
Update in
-
Epigenetic Changes Regulating Epithelial-Mesenchymal Plasticity in Human Trophoblast Differentiation.Cells. 2025 Jun 24;14(13):970. doi: 10.3390/cells14130970. Cells. 2025. PMID: 40643491 Free PMC article.
Abstract
The phenotype of human placental extravillous trophoblast (EVT) at the end of pregnancy reflects both first trimester differentiation from villous cytotrophoblast (CTB) and later gestational changes, including loss of proliferative and invasive capacity. Invasion abnormalities are central to two major placental pathologies, preeclampsia and placenta accreta spectrum, so characterization of the corresponding normal processes is crucial. In this report, our gene expression analysis, using purified human CTB and EVT cells, highlights an epithelial-mesenchymal transition (EMT) mechanism underlying CTB-EVT differentiation and provides a trophoblast-specific EMT signature. In parallel, DNA methylation profiling shows that CTB cells, already hypomethylated relative to non-trophoblast cell lineages, show further genome-wide hypomethylation in the transition to EVT. However, a small subgroup of genes undergoes gains of methylation (GOM) in their regulatory regions or gene bodies, associated with differential mRNA expression (DE). Prominent in this GOM-DE group are genes involved in the EMT, including multiple canonical EMT markers and the EMT-linked transcription factor RUNX1, for which we demonstrate a functional role in modulating the migratory and invasive capacities of JEG3 trophoblast cells. This analysis of DE associated with locus-specific GOM, together with functional studies of an important GOM-DE gene, highlights epigenetically regulated genes and pathways acting in human EVT differentiation and invasion, with implications for obstetric disorders in which these processes are dysregulated.
Keywords: Trophoblast; differentiation; epithelial-mesenchymal transition; methylation.
Conflict of interest statement
Competing interests All authors assert that they have no competing interests to declare
Figures



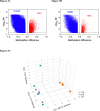


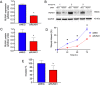
References
-
- Chatterjee A., Macaulay E. C., Rodger E. J., Stockwell P. A., Parry M. F., Roberts H. E., Slatter T. L., Hung N. A., Devenish C. J. and Morison I. M. (2016). Placental Hypomethylation Is More Pronounced in Genomic Loci Devoid of Retroelements. G3 (Bethesda) 6, 1911–1921. 10.1534/g3.116.030379 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
