This is a preprint.
Influenza A defective viral genomes and non-infectious particles are increased by host PI3K inhibition via anti-cancer drug alpelisib
- PMID: 39005364
- PMCID: PMC11245024
- DOI: 10.1101/2024.07.03.601932
Influenza A defective viral genomes and non-infectious particles are increased by host PI3K inhibition via anti-cancer drug alpelisib
Update in
-
Host PI3K inhibition via anti-cancer drug alpelisib influences Influenza A non-infectious particles and deletion-containing viral genomes.Cell Commun Signal. 2025 Dec 19;23(1):543. doi: 10.1186/s12964-025-02598-x. Cell Commun Signal. 2025. PMID: 41419939 Free PMC article.
Abstract
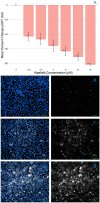
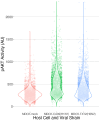
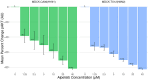
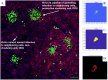
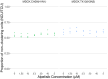
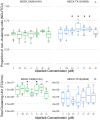
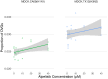
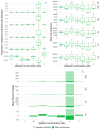
RNA viruses produce abundant defective viral genomes during replication, setting the stage for interactions between viral genomes that alter the course of pathogenesis. Harnessing these interactions to develop antivirals has become a recent goal of intense research focus. Despite decades of research, the mechanisms that regulate the production and interactions of Influenza A defective viral genomes are still unclear. The role of the host is essentially unexplored; specifically, it remains unknown whether host metabolism can influence the formation of defective viral genomes and the particles that house them. To address this question, we manipulated host cell anabolic signaling activity and monitored the production of defective viral genomes and particles by A/H1N1 and A/H3N2 strains, using a combination of single-cell immunofluorescence quantification, third-generation long-read sequencing, and the cluster-forming assay, a method we developed to titer defective and fully-infectious particles simultaneously. Here we show that alpelisib (Piqray), a highly selective inhibitor of mammalian Class 1a phosphoinositide-3 kinase (PI3K) receptors, significantly changed the proportion of defective particles and viral genomes (specifically deletion-containing viral genomes) in a strain-specific manner, under conditions that minimize multiple cycles of replication. Alpelisib pre-treatment of cells led to an increase in defective particles in the A/H3N2 strain, while the A/H1N1 strain showed a decrease in total viral particles. In the same infections, we found that defective viral genomes of polymerase and antigenic segments increased in the A/H1N1 strain, while the total particles decreased suggesting defective interference. We also found that the average deletion size in polymerase complex viral genomes increased in both the A/H3N2 and A/H1N1 strains. The A/H1N1 strain, additionally showed a dose-dependent increase in total number of defective viral genomes. In sum, we provide evidence that host cell metabolism can increase the production of defective viral genomes and particles at an early stage of infection, shifting the makeup of the infection and potential interactions among virions. Given that Influenza A defective viral genomes can inhibit pathogenesis, our study presents a new line of investigation into metabolic states associated with less severe flu infection and the potential induction of these states with metabolic drugs.
Figures

References
-
- Ackermann WW, Klernschmidt E. Concerning the relation of the Krebs cycle to virus propagation. J Biol Chem. 1951. Mar;189(1):421–8. - PubMed
-
- Alnaji FG, Holmes JR, Rendon G, Vera JC, Fields CJ, Martin BE, Brooke CB. Sequencing Framework for the Sensitive Detection and Precise Mapping of Defective Interfering Particle-Associated Deletions across Influenza A and B Viruses. J Virol. 2019. May 15;93(11):e00354–19. doi: 10.1128/JVI.00354-19. - DOI - PMC - PubMed
-
- Al-Saffar NM, Jackson LE, Raynaud FI, Clarke PA, Ramírez de Molina A, Lacal JC, Workman P, Leach MO. The phosphoinositide 3-kinase inhibitor PI-103 downregulates choline kinase alpha leading to phosphocholine and total choline decrease detected by magnetic resonance spectroscopy. Cancer Res. 2010. Jul 1;70(13):5507–17. doi: 10.1158/0008-5472.CAN-09-4476. Epub 2010 Jun 15. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
