This is a preprint.
Co-option of mitochondrial nucleic acid sensing pathways by HSV-1 UL12.5 for reactivation from latent Infection
- PMID: 39005440
- PMCID: PMC11245091
- DOI: 10.1101/2024.07.06.601241
Co-option of mitochondrial nucleic acid sensing pathways by HSV-1 UL12.5 for reactivation from latent Infection
Update in
-
Co-option of mitochondrial nucleic acid-sensing pathways by HSV-1 UL12.5 for reactivation from latent infection.Proc Natl Acad Sci U S A. 2025 Jan 28;122(4):e2413965122. doi: 10.1073/pnas.2413965122. Epub 2025 Jan 24. Proc Natl Acad Sci U S A. 2025. PMID: 39854226 Free PMC article.
Abstract
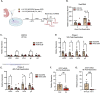
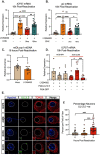
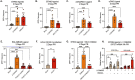
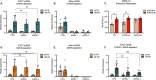
Although viruses subvert innate immune pathways for their replication, there is evidence they can also co-opt anti-viral responses for their benefit. The ubiquitous human pathogen, Herpes Simplex Virus-1 (HSV-1), encodes a protein (UL12.5) that induces the release of mitochondrial nucleic acid into the cytosol, which activates immune sensing pathways and reduces productive replication in non-neuronal cells. HSV-1 establishes latency in neurons and can reactivate to cause disease. We found that UL12.5 is required for HSV-1 reactivation in neurons and acts to directly promote viral lytic gene expression during initial exit from latency. Further, the direct activation of innate immune sensing pathways triggered HSV reactivation and compensated for a lack of UL12.5. Finally, we found that the induction of HSV-1 lytic genes during reactivation required intact RNA and DNA sensing pathways, demonstrating that HSV-1 can both respond to and active antiviral nucleic acid sensing pathways to reactivate from a latent infection.
Figures


References
-
- Aguirre S., Luthra P., Sanchez-Aparicio M.T., Maestre A.M., Patel J., Lamothe F., Fredericks A.C., Tripathi S., Zhu T., Pintado-Silva J., et al. (2017). Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat Microbiol 2, 17037. 10.1038/nmicrobiol.2017.37. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
