Insights on the conformation and appropriate drug-target sites on retinal IMPDH1 using the 604-aa isoform lacking the C-terminal extension
- PMID: 39005562
- PMCID: PMC11246115
- DOI: 10.4103/1735-5362.389951
Insights on the conformation and appropriate drug-target sites on retinal IMPDH1 using the 604-aa isoform lacking the C-terminal extension
Abstract
Background and purpose: Retinitis pigmentosa (RP) accounts for 2 percent of global cases of blindness. The RP10 form of the disease results from mutations in isoform 1 of inosine 5'-monophosphate dehydrogenase (IMPDH1), the rate-limiting enzyme in the de novo purine nucleotide synthesis pathway. Retinal photoreceptors contain specific isoforms of IMPDH1 characterized by terminal extensions. Considering previously reported significantly varied kinetics among retinal isoforms, the current research aimed to investigate possible structural explanations and suitable functional sites for the pharmaceutical targeting of IMPDH1 in RP.
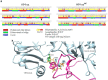
Experimental approach: A recombinant 604-aa IMPDH1 isoform lacking the carboxyl-terminal peptide was produced and underwent proteolytic digestion with α-chymotrypsin. Dimer models of wild type and engineered 604-aa isoform were subjected to molecular dynamics simulation.
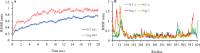
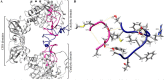
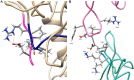
Findings/results: The IMPDH1 retinal isoform lacking C-terminal peptide was shown to tend to have more rapid proteolysis (~16% digestion in the first two minutes). Our computational data predicted the potential of the amino-terminal peptide to induce spontaneous inhibition of IMPDH1 by forming a novel helix in a GTP binding site. On the other hand, the C-terminal peptide might block the probable inhibitory role of the N-terminal extension.
Conclusion and implications: According to the findings, augmenting IMPDH1 activity by suppressing its filamentation is suggested as a suitable strategy to compensate for its disrupted activity in RP. This needs specific small molecule inhibitors to target the filament assembly interface of the enzyme.
Keywords: Inosine monophosphate dehydrogenase; Molecular dynamics simulation; Proteolytic digestion; Retinal isoforms; Retinitis pigmentosa.
Copyright: © 2023 Research in Pharmaceutical Sciences.
Conflict of interest statement
The authors declared no conflict of interest in this study.
Figures

References
-
- Majd N, Sumita K, Yoshino H, Chen D, Terakawa J, Daikoku T, et al. A review of the potential utility of mycophenolate mofetil as a cancer therapeutic. J Cancer Res. 2014;2014(6):1–12. DOI: 10.1155/2014/423401.
-
- Baykov AA, Tuominen HK, Lahti R. The CBS domain: a protein module with an emerging prominent role in regulation. ACS Chem Biol. 2011;6(11):1156–1163. DOI: 10.1021/cb200231c. - PubMed
LinkOut - more resources
Full Text Sources
