Artificial Intelligence-Clinical Decision Support System in Infectious Disease Control: Combatting Multidrug-Resistant Klebsiella pneumoniae with Machine Learning
- PMID: 39005853
- PMCID: PMC11246630
- DOI: 10.2147/IDR.S470821
Artificial Intelligence-Clinical Decision Support System in Infectious Disease Control: Combatting Multidrug-Resistant Klebsiella pneumoniae with Machine Learning
Abstract
Purpose: The World Health Organization has identified Klebsiella pneumoniae (KP) as a significant threat to global public health. The rising threat of carbapenem-resistant Klebsiella pneumoniae (CRKP) leads to prolonged hospital stays and higher medical costs, necessitating faster diagnostic methods. Traditional antibiotic susceptibility testing (AST) methods demand at least 4 days, requiring 3 days on average for culturing and isolating the bacteria and identifying the species using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), plus an extra day for interpreting AST results. This lengthy process makes traditional methods too slow for urgent clinical situations requiring rapid decision-making, potentially hindering prompt treatment decisions, especially for fast-spreading infections such as those caused by CRKP. This research leverages a cutting-edge diagnostic method that utilizes an artificial intelligence-clinical decision support system (AI-CDSS). It incorporates machine learning algorithms for the swift and precise detection of carbapenem-resistant and colistin-resistant strains.
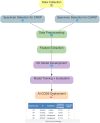
Patients and methods: We selected 4307 KP samples out of a total of 52,827 bacterial samples due to concerns about multi-drug resistance using MALDI-TOF MS and Vitek-2 systems for AST. It involved thorough data preprocessing, feature extraction, and machine learning model training fine-tuned with GridSearchCV and 5-fold cross-validation, resulting in high predictive accuracy, as demonstrated by the receiver operating characteristic and area under the curve (AUC) scores, laying the groundwork for our AI-CDSS.
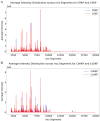
Results: MALDI-TOF MS analysis revealed distinct intensity profiles differentiating CRKP and susceptible strains, as well as colistin-resistant Klebsiella pneumoniae (CoRKP) and susceptible strains. The Random Forest Classifier demonstrated superior discriminatory power, with an AUC of 0.96 for detecting CRKP and 0.98 for detecting CoRKP.
Conclusion: Integrating MALDI-TOF MS with machine learning in an AI-CDSS has greatly expedited the detection of KP resistance by approximately 1 day. This system offers timely guidance, potentially enhancing clinical decision-making and improving treatment outcomes for KP infections.
Keywords: MALDI-TOF MS; antibiotic stewardship; carbapenem; colistin; diagnostic accuracy.
© 2024 Jian et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures



References
-
- World Health Organization. WHO publishes list of bacteria for which new antibiotics are urgently needed; 2017. Available from: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-.... Accessed April 16, 2024.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

