Atopic Dermatitis Complicated by Recurrent Eczema Herpeticum Is Characterized by Multiple, Concurrent Epidermal Inflammatory Endotypes
- PMID: 39006317
- PMCID: PMC11239700
- DOI: 10.1016/j.xjidi.2024.100279
Atopic Dermatitis Complicated by Recurrent Eczema Herpeticum Is Characterized by Multiple, Concurrent Epidermal Inflammatory Endotypes
Abstract
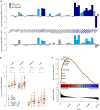
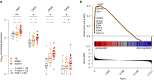
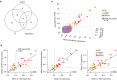
A subgroup of patients with atopic dermatitis (AD) suffers from recurrent, disseminated herpes simplex virus skin infection, termed eczema herpeticum. To determine the transcriptional mechanisms of the skin and immune system pathobiology that underlie development of AD with eczema herpeticum (ADEH), we performed RNA-sequencing analysis of nonlesional skin (epidermis, dermis) from AD patients with and without a history of ADEH (ADEH+, n = 15; ADEH-, n = 13) along with healthy controls (n = 15). We also performed RNA sequencing on participants' plasmacytoid dendritic cells infected in vitro with herpes simplex virus 1. ADEH+ patients exhibited dysregulated gene expression, limited in the dermis (14 differentially expressed genes) and more widespread in the epidermis (129 differentially expressed genes). ADEH+-upregulated epidermal differentially expressed genes were enriched in type 2 cytokine (IL4R , CCL22, CRLF2, IL7R), interferon (CXCL10, ICAM1, IFI44, IRF7), and IL-36γ (IL36G) inflammatory gene pathways. All ADEH+ participants exhibited type 2 cytokine and inteferon endotypes, and 87% were IL36G-high. In contrast, these endotypes were more variably expressed among ADEH- participants. ADEH+ skin also had dysregulated epidermal differentiation complex gene expression of the late-cornified envelope, S100A, and small proline-rich gene families, which are involved in skin barrier function and antimicrobial activities. Plasmacytoid dendritic cell transcriptional responses to herpes simplex virus 1 infection were unaltered by ADEH status. The study concluded that the pathobiology underlying ADEH+ risk is associated with a unique, multifaceted epidermal inflammation that accompanies dysregulation of epidermal differentiation complex genes. These findings will help direct future studies that define how these inflammatory patterns may drive risk of eczema herpeticum in AD.
Keywords: AD; ADEH; HSV; RNA sequencing; Skin disease.
© 2024 The Authors.
Figures



Update of
-
Atopic dermatitis complicated by recurrent eczema herpeticum is characterized by multiple, concurrent epidermal inflammatory endotypes.bioRxiv [Preprint]. 2023 Feb 28:2023.02.27.530316. doi: 10.1101/2023.02.27.530316. bioRxiv. 2023. Update in: JID Innov. 2024 Apr 02;4(4):100279. doi: 10.1016/j.xjidi.2024.100279. PMID: 36909594 Free PMC article. Updated. Preprint.
References
-
- Abtin A., Eckhart L., Gläser R., Gmeiner R., Mildner M., Tschachler E. The antimicrobial heterodimer S100A8/S100A9 (calprotectin) is upregulated by bacterial flagellin in human epidermal keratinocytes. J Invest Dermatol. 2010;130:2423–2430. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

