Single-Cell Analysis Identifies Distinct Populations of Cytotoxic CD4+ T Cells Linked to the Therapeutic Efficacy of Immune Checkpoint Inhibitors in Metastatic Renal Cell Carcinoma
- PMID: 39006494
- PMCID: PMC11246657
- DOI: 10.2147/JIR.S457570
Single-Cell Analysis Identifies Distinct Populations of Cytotoxic CD4+ T Cells Linked to the Therapeutic Efficacy of Immune Checkpoint Inhibitors in Metastatic Renal Cell Carcinoma
Abstract
Background: The involvement of cytotoxic CD4+ T cells (CD4+ CTLs) and their potential role in dictating the response to immune checkpoint inhibitors (ICIs) in patients with metastatic renal cell carcinoma (mRCC) remains an unexplored area of research.
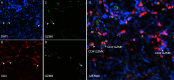
Methods: Utilizing single-cell RNA sequencing, we analyzed the immunophenotype and expression patterns of CD4+ T lymphocyte subtypes in mRCC patients, followed by preliminary validation via multi-immunofluorescent staining. In addition, we obtained a comprehensive immunotherapy dataset encompassing single-cell RNA sequencing datasets and bulk RNA-seq cohorts from the European Genome-Phenome Archive and ArrayExpress database. Utilizing the CIBERSORTx deconvolution algorithms, we derived a signature score for CD4+ CTLs from the bulk-RNA-seq datasets of the CheckMate 009/025 clinical trials.
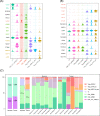
Results: Single-cell analysis of CD4+ T lymphocytes in mRCC reveals several cancer-specific states, including diverse phenotypes of regulatory T cells. Remarkably, we observe that CD4+ CTLs cells constitute a substantial proportion of all CD4+ T lymphocyte sub-clusters in mRCC patients, highlighting their potential significance in the disease. Furthermore, within mRCC patients, we identify two distinct cytotoxic states of CD4+ T cells: CD4+GZMK+ T cells, which exhibit a weaker cytotoxic potential, and CD4+GZMB+ T cells, which demonstrate robust cytotoxic activity. Both regulatory T cells and CD4+ CTLs originate from proliferating CD4+ T cells within mRCC tissues. Intriguingly, our trajectory analysis indicates that the weakly cytotoxic CD4+GZMK+ T cells differentiate from their more cytotoxic CD4+GZMB+ counterparts. In comparing patients with lower CD4+ CTLs levels to those with higher CD4+ CTLs abundance in the CheckMate 009 and 25 immunotherapy cohorts, the latter group exhibited significantly improved OS and PFS probability.
Conclusion: Our study underscores the pivotal role that intratumoral CD4+ CTLs may play in bolstering anti-tumor immunity, suggesting their potential as a promising biomarker for predicting response to ICIs in patients with mRCC.
Keywords: cytotoxic CD4+ T cells; granzyme K and granzyme B; immune checkpoint inhibitors; renal cell carcinoma; single-cell analysis.
© 2024 Yang et al.
Conflict of interest statement
The authors declare that they have no competing interests in this work.
Figures






References
LinkOut - more resources
Full Text Sources
Research Materials

