Identifying regulatory elements and their RNA-binding proteins in the 3' untranslated regions of papillomavirus late mRNAs
- PMID: 39006509
- PMCID: PMC11240274
- DOI: 10.3892/br.2024.1813
Identifying regulatory elements and their RNA-binding proteins in the 3' untranslated regions of papillomavirus late mRNAs
Abstract
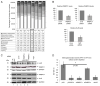
Human papillomaviruses (HPVs) infect cutaneous and mucosal epithelia to cause benign (warts) and malignant lesions (e.g. cervical cancer). Bovine papillomaviruses (BPVs) infect fibroblasts to cause fibropapillomas but can also infect cutaneous epithelial cells. For HPV-1, -16, -31 and BPV-1, cis-acting RNA elements in the late 3' untranslated region (3'UTR) control expression of virus proteins by binding host cell proteins. The present study compared the effects on gene expression of the cis-acting elements of seven PV late 3'UTRs (HPV-6b, -11, -16, -31 and BPV-1, -3 and -4) representing a range of different genera and species and pathological properties. pSV-beta-galactosidase reporter plasmids containing the late 3'UTRs from seven PVs were transiently transfected into cervical adenocarcinoma HeLa cells, and reporter gene expression quantified by reverse transcription-quantitative PCR and a beta-galactosidase assay. All elements inhibited gene expression in keratinocytes. Cancer-related types HPV-16 and -31, had the greatest inhibitory activity whereas the lowest inhibition was found in the non-cancer related types, BPV-3 and HPV-11. Using RBPmap version 1.1, bioinformatics predictions of factors binding the elements identified proteins which function mainly in mRNA splicing. Markedly, in terms of protein binding motifs, BPV late 3'UTR elements were similar to those of HPV-1a but not to other HPVs. Using HPV-1a as a model and siRNA depletion, the bioinformatics predictions were tested and it was found that PABPC4 was responsible for some of the 3'UTR repressive activity. The data revealed candidate proteins that could control PV late gene expression.
Keywords: 3'untranslated region; RNA regulatory element; RNA-binding protein motifs; late gene expression; papillomavirus.
Copyright: © 2024 Iamborwornkun et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
LinkOut - more resources
Full Text Sources
Research Materials
