A hot and cold tumor‑related prognostic signature for stage II colorectal cancer
- PMID: 39006949
- PMCID: PMC11240279
- DOI: 10.3892/ol.2024.14552
A hot and cold tumor‑related prognostic signature for stage II colorectal cancer
Abstract
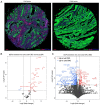
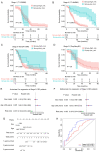
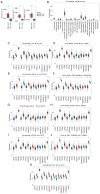
Globally, colorectal cancer (CRC) is one of the most lethal and prevalent malignancies. Based on the presence of immune cell infiltration in the tumor microenvironment, CRC can be divided into immunologically 'hot' or 'cold' tumors, which in turn leads to the differential efficacy of immunotherapy. However, the immune characteristics of hot and cold CRC tumors remain largely elusive, prompting further investigation of their properties regarding the tumor microenvironment. In the present study, a predictive model was developed based on the differential expression of proteins between cold and hot CRC tumors. First, the differentially expressed proteins (DEPs) were identified using digital spatial profiling and mass spectrometry-based proteomics analysis, and the pathway features of the DEPs were analyzed using functional enrichment analysis. A novel eight-gene signature prognostic risk model was developed (IDO1, MAT1A, NPEPL1, NT5C, PTGR2, RPL29, TMEM126A and TUBB4B), which was validated using data obtained from The Cancer Genome Atlas. The results revealed that the risk score of the eight-gene signature acted as an independent prognostic indicator in patients with stage II CRC (T3-4N0M0). It was also found that a high-risk score in the eight-gene signature was associated with high immune cell infiltration in patients with CRC. Taken together, these findings revealed some of the differential immune characteristics of hot and cold CRC tumors, and an eight-gene signature prognostic risk model was developed, which may serve as an independent prognostic indicator for patients with stage II CRC (T3-4N0M0).
Keywords: colorectal cancer; hot and cold tumor; prognosis; tumor microenvironment.
Copyright: © 2024 Zhou et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
