Diagnostic model for hepatocellular carcinoma using small extracellular vesicle-propagated miRNA signatures
- PMID: 39006969
- PMCID: PMC11239443
- DOI: 10.3389/fmolb.2024.1419093
Diagnostic model for hepatocellular carcinoma using small extracellular vesicle-propagated miRNA signatures
Abstract
Background: Hepatocellular carcinoma (HCC) is the most common type of liver cancer. Small extracellular vesicles (sEVs) are bilayer lipid membrane vesicles containing RNA that exhibit promising diagnostic and prognostic potential as cancer biomarkers.
Aims: To establish a miRNA panel from peripheral blood for use as a noninvasive biomarker for the diagnosis of HCC.
Methods: sEVs obtained from plasma were profiled using high-throughput sequencing. The identified differential miRNA expression patterns were subsequently validated using quantitative real-time polymerase chain reaction analysis.
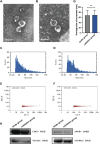
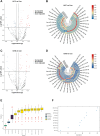
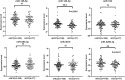
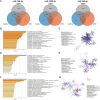
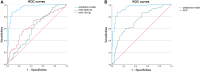
Results: The random forest method identified ten distinct miRNAs distinguishing HCC plasma from non-HCC plasma. During validation, miR-140-3p (p = 0.0001) and miR-3200-3p (p = 0.0017) exhibited significant downregulation. Enrichment analysis uncovered a notable correlation between the target genes of these miRNAs and cancer development. Utilizing logistic regression, we developed a diagnostic model incorporating these validated miRNAs. Receiver operating characteristic (ROC) curve analysis revealed an area under the curve (AUC) of 0.951, with a sensitivity of 90.1% and specificity of 87.8%.
Conclusion: These aberrantly expressed miRNAs delivered by sEVs potentially contribute to HCC pathology and may serve as diagnostic biomarkers for HCC.
Keywords: biomarker; cancer; hepatocellular carcinoma; microRNA; small extracellular vesicle.
Copyright © 2024 Luo, Jiao, Guo, Chen, Wang, Wen, Song, Chen, Zhou and Song.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources

