Molecular analysis of primary and metastatic sites in patients with renal cell carcinoma
- PMID: 39007269
- PMCID: PMC11245151
- DOI: 10.1172/JCI176230
Molecular analysis of primary and metastatic sites in patients with renal cell carcinoma
Abstract
BACKGROUNDMetastases are the hallmark of lethal cancer, though underlying mechanisms that drive metastatic spread to specific organs remain poorly understood. Renal cell carcinoma (RCC) is known to have distinct sites of metastases, with lung, bone, liver, and lymph nodes being more common than brain, gastrointestinal tract, and endocrine glands. Previous studies have shown varying clinical behavior and prognosis associated with the site of metastatic spread; however, little is known about the molecular underpinnings that contribute to the differential outcomes observed by the site of metastasis.METHODSWe analyzed primary renal tumors and tumors derived from metastatic sites to comprehensively characterize genomic and transcriptomic features of tumor cells as well as to evaluate the tumor microenvironment at both sites.RESULTSWe included a total of 657 tumor samples (340 from the primary site [kidney] and 317 from various sites of metastasis). We show distinct genomic alterations, transcriptomic signatures, and immune and stromal tumor microenvironments across metastatic sites in a large cohort of patients with RCC.CONCLUSIONWe demonstrate significant heterogeneity among primary tumors and metastatic sites and elucidate the complex interplay between tumor cells and the extrinsic tumor microenvironment that is vital for developing effective anticancer therapies.
Keywords: Cancer; Oncology.
Conflict of interest statement
Figures
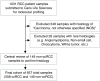
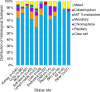




References
-
- SEER Cancer Stat Facts: Cancer of Any Site. National Cancer Institute. https://seer.cancer.gov/statfacts/html/all.html.
-
- Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989;8(2):98–101. - PubMed

