Detection and characterization of H5N1 HPAIV in environmental samples from a dairy farm
- PMID: 39008139
- PMCID: PMC11686969
- DOI: 10.1007/s11262-024-02085-4
Detection and characterization of H5N1 HPAIV in environmental samples from a dairy farm
Abstract
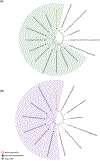
The recent expansion of HPAIV H5N1 infections in terrestrial mammals in the Americas, most recently including the outbreak in dairy cattle, emphasizes the critical need for better epidemiological monitoring of zoonotic diseases. In this work, we detected, isolated, and characterized the HPAIV H5N1 from environmental swab samples collected from a dairy farm in the state of Kansas, USA. Genomic sequencing of these samples uncovered two distinctive substitutions in the PB2 (E249G) and NS1 (R21Q) genes which are rare and absent in recent 2024 isolates of H5N1 circulating in the mammalian and avian species. Additionally, approximately 1.7% of the sequence reads indicated a PB2 (E627K) substitution, commonly associated with virus adaptation to mammalian hosts. Phylogenetic analyses of the PB2 and NS genes demonstrated more genetic identity between this environmental isolate and the 2024 human isolate (A/Texas/37/2024) of H5N1. Conversely, HA and NA gene analyses revealed a closer relationship between our isolate and those found in other dairy cattle with almost 100% identity, sharing a common phylogenetic subtree. These findings underscore the rapid evolutionary progression of HPAIV H5N1 among dairy cattle and reinforces the need for more epidemiological monitoring which can be done using environmental sampling.
Keywords: Bovine; Environmental; H5N1; Highly pathogenic avian influenza virus; Mammalian.
© 2024. The Author(s), under exclusive licence to Springer Science+Business Media, LLC, part of Springer Nature.
Figures
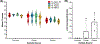



Similar articles
-
A One Health Investigation into H5N1 Avian Influenza Virus Epizootics on Two Dairy Farms.Clin Infect Dis. 2025 Feb 24;80(2):331-338. doi: 10.1093/cid/ciae576. Clin Infect Dis. 2025. PMID: 39658318
-
Avian Influenza A(H5N1) Virus among Dairy Cattle, Texas, USA.Emerg Infect Dis. 2024 Jul;30(7):1425-1429. doi: 10.3201/eid3007.240717. Epub 2024 Jun 7. Emerg Infect Dis. 2024. PMID: 38848249 Free PMC article.
-
Spillover of highly pathogenic avian influenza H5N1 virus to dairy cattle.Nature. 2024 Oct;634(8034):669-676. doi: 10.1038/s41586-024-07849-4. Epub 2024 Jul 25. Nature. 2024. PMID: 39053575 Free PMC article.
-
An Update on Highly Pathogenic Avian Influenza A(H5N1) Virus, Clade 2.3.4.4b.J Infect Dis. 2024 Sep 23;230(3):533-542. doi: 10.1093/infdis/jiae379. J Infect Dis. 2024. PMID: 39283944 Review.
-
mGem: Transmission and exposure risks of dairy cow H5N1 influenza virus.mBio. 2025 Mar 12;16(3):e0294424. doi: 10.1128/mbio.02944-24. Epub 2025 Feb 11. mBio. 2025. PMID: 39932310 Free PMC article. Review.
Cited by
-
Risk posed by the HPAI virus H5N1, Eurasian lineage goose/Guangdong clade 2.3.4.4b. genotype B3.13, currently circulating in the US.EFSA J. 2025 Jul 3;23(7):e9508. doi: 10.2903/j.efsa.2025.9508. eCollection 2025 Jul. EFSA J. 2025. PMID: 40611987 Free PMC article.
-
Outcome of H5N1 clade 2.3.4.4b virus infection in calves and lactating cows.bioRxiv [Preprint]. 2024 Aug 9:2024.08.09.607272. doi: 10.1101/2024.08.09.607272. bioRxiv. 2024. PMID: 39149352 Free PMC article. Preprint.
-
The emergence of highly pathogenic avian influenza H5N1 in dairy cattle: implications for public health, animal health, and pandemic preparedness.Eur J Clin Microbiol Infect Dis. 2025 Aug;44(8):1817-1833. doi: 10.1007/s10096-025-05147-z. Epub 2025 May 14. Eur J Clin Microbiol Infect Dis. 2025. PMID: 40369349 Free PMC article. Review.
-
Avian Influenza Virus A(H5Nx) and Prepandemic Candidate Vaccines: State of the Art.Int J Mol Sci. 2024 Aug 5;25(15):8550. doi: 10.3390/ijms25158550. Int J Mol Sci. 2024. PMID: 39126117 Free PMC article. Review.
-
Susceptibility and shedding in Mx1+ and Mx1- female mice experimentally infected with dairy cattle A(H5N1) influenza viruses.EBioMedicine. 2025 Jul 8;118:105842. doi: 10.1016/j.ebiom.2025.105842. Online ahead of print. EBioMedicine. 2025. PMID: 40633142 Free PMC article.
References
-
- Claas EC et al. (1998) Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351:472–477 - PubMed
-
- Senne DA et al. (1996) Survey of the hemagglutinin (HA) cleavage site sequence of H5 and H7 Avian Influenza viruses: amino acid sequence at the HA cleavage site as a marker of pathogenicity potential. Avian Dis. 40:425–437 - PubMed
-
- CDC (2024) Current H5N1 bird flu situation in dairy cows. Avian Influenza (Bird Flu) https://www.cdc.gov/bird-flu/situation-summary/mammals.html.
-
- Avian influenza virus type A (H5N1) in U.S. dairy cattle | American Veterinary Medical Association. https://www.avma.org/resources-tools/animal-health-and-welfare/animal-he....
-
- Highly Pathogenic Avian Influenza (HPAI) Detections in Live-stock | Animal and Plant Health Inspection Service. https://www.aphis.usda.gov/livestock-poultry-disease/avian/avian-influen....
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

