Genomic characterization of highly pathogenic avian influenza A H5N1 virus newly emerged in dairy cattle
- PMID: 39008278
- PMCID: PMC11271078
- DOI: 10.1080/22221751.2024.2380421
Genomic characterization of highly pathogenic avian influenza A H5N1 virus newly emerged in dairy cattle
Abstract
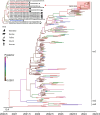
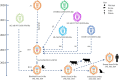
In March 2024, the emergence of highly pathogenic avian influenza (HPAI) A (H5N1) infections in dairy cattle was detected in the United Sates for the first time. We genetically characterize HPAI viruses from dairy cattle showing an abrupt drop in milk production, as well as from two cats, six wild birds, and one skunk. They share nearly identical genome sequences, forming a new genotype B3.13 within the 2.3.4.4b clade. B3.13 viruses underwent two reassortment events since 2023 and exhibit critical mutations in HA, M1, and NS genes but lack critical mutations in PB2 and PB1 genes, which enhance virulence or adaptation to mammals. The PB2 E627 K mutation in a human case associated with cattle underscores the potential for rapid evolution post infection, highlighting the need for continued surveillance to monitor public health threats.
Keywords: H5N1; Highly pathogenic avian influenza (HPAI); clade 2.3.4.4b; dairy cattle; genome sequence; reassortment events.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
References
-
- Youk S, Torchetti MK, Lantz K, et al. H5N1 highly pathogenic avian influenza clade 2.3. 4.4 b in wild and domestic birds: Introductions into the United States and reassortments, December 2021–April 2022. Virology. 2023;587:109860. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
