A genome assembly of the American black bear, Ursus americanus, from California
- PMID: 39008331
- PMCID: PMC11334205
- DOI: 10.1093/jhered/esae037
A genome assembly of the American black bear, Ursus americanus, from California
Abstract
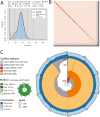
The American black bear, Ursus americanus, is a widespread and ecologically important species in North America. In California, the black bear plays an important role in a variety of ecosystems and serves as an important species for recreational hunting. While research suggests that the populations in California are currently healthy, continued monitoring is critical, with genomic analyses providing an important surveillance tool. Here we report a high-quality, near chromosome-level genome assembly from a U. americanus sample from California. The primary assembly has a total length of 2.5 Gb contained in 316 scaffolds, a contig N50 of 58.9 Mb, a scaffold N50 of 67.6 Mb, and a BUSCO completeness score of 96%. This U. americanus genome assembly will provide an important resource for the targeted management of black bear populations in California, with the goal of achieving an appropriate balance between the recreational value of black bears and the maintenance of viable populations. The high quality of this genome assembly will also make it a valuable resource for comparative genomic analyses among black bear populations and among bear species.
Keywords: CCGP; California Conservation Genomics Project; Conservation Genomics; wildlife management.
© The American Genetic Association. 2024.
Figures

References
-
- California Department of Fish and Game. Black Bear Management Plan. 1998. [Accessed 14 February 2024].https://nrm.dfg.ca.gov/FileHandler.ashx?DocumentID=82769&inline
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

