Sequencing of Kaposi's Sarcoma Herpesvirus (KSHV) genomes from persons of diverse ethnicities and provenances with KSHV-associated diseases demonstrate multiple infections, novel polymorphisms, and low intra-host variance
- PMID: 39008527
- PMCID: PMC11271956
- DOI: 10.1371/journal.ppat.1012338
Sequencing of Kaposi's Sarcoma Herpesvirus (KSHV) genomes from persons of diverse ethnicities and provenances with KSHV-associated diseases demonstrate multiple infections, novel polymorphisms, and low intra-host variance
Abstract
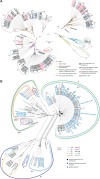
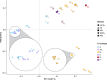
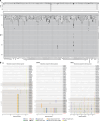
Recently published near full-length KSHV genomes from a Cameroon Kaposi sarcoma case-control study showed strong evidence of viral recombination and mixed infections, but no sequence variations associated with disease. Using the same methodology, an additional 102 KSHV genomes from 76 individuals with KSHV-associated diseases have been sequenced. Diagnoses comprise all KSHV-associated diseases (KAD): Kaposi sarcoma (KS), primary effusion lymphoma (PEL), KSHV-associated large cell lymphoma (KSHV-LCL), a type of multicentric Castleman disease (KSHV-MCD), and KSHV inflammatory cytokine syndrome (KICS). Participants originated from 22 different countries, providing the opportunity to obtain new near full-length sequences of a wide diversity of KSHV genomes. These include near full-length sequence of genomes with KSHV K1 subtypes A, B, C, and F as well as subtype E, for which no full sequence was previously available. High levels of recombination were observed. Fourteen individuals (18%) showed evidence of infection with multiple KSHV variants (from two to four unique genomes). Twenty-six comparisons of sequences, obtained from various sampling sites including PBMC, tissue biopsies, oral fluids, and effusions in the same participants, identified near complete genome conservation between different biological compartments. Polymorphisms were identified in coding and non-coding regions, including indels in the K3 and K15 genes and sequence inversions here reported for the first time. One such polymorphism in KSHV ORF46, specific to the KSHV K1 subtype E2, encoded a mutation in the leucine loop extension of the uracil DNA glycosylase that results in alteration of biochemical functions of this protein. This confirms that KSHV sequence variations can have functional consequences warranting further investigation. This study represents the largest and most diverse analysis of KSHV genome sequences to date among individuals with KAD and provides important new information on global KSHV genomics.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
R. Yarchoan reports receiving research support from Celgene (now Bristol Myers Squibb), CTI BioPharma (a Sobi A.B. Company), PDS Biotech, and Janssen Pharmaceuticals through CRADAs with the NCI. Dr. Yarchoan also reports receiving drugs for clinical trials from Merck, EMD-Serano, and Eli Lilly and preclinical material from Lentigen Technology through CRADAs or MTAs with the NCI. R. Yarchoan and T.S.Uldrick are co-inventors on US Patent 10,001,483 entitled "Methods for the treatment of Kaposi’s sarcoma or KSHV-induced lymphoma using immunomodulatory compounds and uses of biomarkers." An immediate family member of R. Yarchoan is a co-inventor on patents or patent applications related to internalization of target receptors, epigenetic analysis, and ephrin tyrosine kinase inhibitors. All rights, title, and interest to these patents have been assigned to the U.S. Department of Health and Human Services; the government conveys a portion of the royalties it receives to its employee inventors under the Federal Technology Transfer Act of 1986 (P.L. 99-502). Thomas Uldrick is also co-inventor on U.S. patent application no. 18/310,649, KSHV ONCOPROTEIN ANTIGENS AND EPITOPES FOR EXPANDING ANTIGEN-SPECIFIC T CELLS. Drs. Kathryn Lurain and Ramya Ramaswami receive research support from Bristol Myers Squibb, Merck, EMD-Serono, Eli Lilly, Lentigen, CTI BioPharma, and Janssen through CRADAs with the NCI. No potential conflicts of interest were disclosed by the other authors.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

