Conformational dynamics underlying atypical chemokine receptor 3 activation
- PMID: 39008676
- PMCID: PMC11287255
- DOI: 10.1073/pnas.2404000121
Conformational dynamics underlying atypical chemokine receptor 3 activation
Abstract
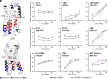
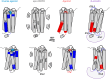
Atypical Chemokine Receptor 3 (ACKR3) belongs to the G protein-coupled receptor family but it does not signal through G proteins. The structural properties that govern the functional selectivity and the conformational dynamics of ACKR3 activation are poorly understood. Here, we combined hydrogen/deuterium exchange mass spectrometry, site-directed mutagenesis, and molecular dynamics simulations to examine the binding mode and mechanism of action of ACKR3 ligands of different efficacies. Our results show that activation or inhibition of ACKR3 is governed by intracellular conformational changes of helix 6, intracellular loop 2, and helix 7, while the DRY motif becomes protected during both processes. Moreover, we identified the binding sites and the allosteric modulation of ACKR3 upon β-arrestin 1 binding. In summary, this study highlights the structure-function relationship of small ligands, the binding mode of β-arrestin 1, the activation dynamics, and the atypical dynamic features in ACKR3 that may contribute to its inability to activate G proteins.
Keywords: ACKR3; GPCR conformational dynamics; HDX-MS; MD simulations.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



References
-
- Griffith J. W., Sokol C. L., Luster A. D., Chemokines and chemokine receptors: Positioning cells for host defense and immunity. Annu. Rev. Immunol. 32, 659–702 (2014). - PubMed
-
- Balabanian K., et al. , The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J. Biol. Chem. 280, 35760–35766 (2005). - PubMed
-
- Smit M. J., et al. , The CXCL12/CXCR4/ACKR3 axis in the tumor microenvironment: Signaling, crosstalk, and therapeutic targeting. Annu. Rev. Pharmacol. Toxicol. 61, 541–563 (2021). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

