Novel insights into paclitaxel's role on tumor-associated macrophages in enhancing PD-1 blockade in breast cancer treatment
- PMID: 39009452
- PMCID: PMC11253755
- DOI: 10.1136/jitc-2024-008864
Novel insights into paclitaxel's role on tumor-associated macrophages in enhancing PD-1 blockade in breast cancer treatment
Abstract
Background: Triple-negative breast cancer (TNBC) poses unique challenges due to its complex nature and the need for more effective treatments. Recent studies showed encouraging outcomes from combining paclitaxel (PTX) with programmed cell death protein-1 (PD-1) blockade in treating TNBC, although the exact mechanisms behind the improved results are unclear.
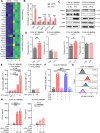
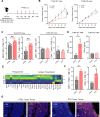
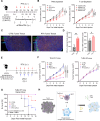
Methods: We employed an integrated approach, analyzing spatial transcriptomics and single-cell RNA sequencing data from TNBC patients to understand why the combination of PTX and PD-1 blockade showed better response in TNBC patients. We focused on toll-like receptor 4 (TLR4), a receptor of PTX, and its role in modulating the cross-presentation signaling pathways in tumor-associated macrophages (TAMs) within the tumor microenvironment. Leveraging insights obtained from patient-derived data, we conducted in vitro experiments using immunosuppressive bone marrow-derived macrophages (iBMDMs) to validate if PTX could augment the cross-presentation and phagocytosis activities. Subsequently, we extended our study to an in vivo murine model of TNBC to ascertain the effects of PTX on the cross-presentation capabilities of TAMs and its downstream impact on CD8+ T cell-mediated immune responses.
Results: Data analysis from TNBC patients revealed that the activation of TLR4 and cross-presentation signaling pathways are crucial for the antitumor efficacy of PTX. In vitro studies showed that PTX treatment enhances the cross-presentation ability of iBMDMs. In vivo experiments demonstrated that PTX activates TLR4-dependent cross-presentation in TAMs, improving CD8+ T cell-mediated antitumor responses. The efficacy of PTX in promoting antitumor immunity was elicited when combined with PD-1 blockade, suggesting a complementary interaction.
Conclusions: This study reveals how PTX boosts the effectiveness of PD-1 inhibitors in treating TNBC. We found that PTX activates TLR4 signaling in TAMs. This activation enhances their ability to present antigens, thereby boosting CD8+ T cell antitumor responses. These findings not only shed light on PTX's immunomodulatory role in TNBC but also underscore the potential of targeting TAMs' antigen presentation capabilities in immunotherapy approaches.
Keywords: Breast Cancer; Combination therapy; Immune Checkpoint Inhibitor; Macrophage; Toll-like receptor - TLR.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: G-HN and I-SK are the cofounders, and YC, EK, YKK, SK, and JK are employees of SHIFTIBIO. HC is a cofounder, and DL is an employee of Portrai. The other authors declare no conflicts of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
