Single-cell transcriptomics analysis of bullous pemphigoid unveils immune-stromal crosstalk in type 2 inflammatory disease
- PMID: 39009587
- PMCID: PMC11251189
- DOI: 10.1038/s41467-024-50283-3
Single-cell transcriptomics analysis of bullous pemphigoid unveils immune-stromal crosstalk in type 2 inflammatory disease
Abstract
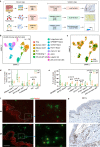
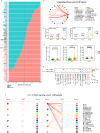
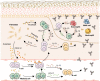
Bullous pemphigoid (BP) is a type 2 inflammation- and immunity-driven skin disease, yet a comprehensive understanding of the immune landscape, particularly immune-stromal crosstalk in BP, remains elusive. Herein, using single-cell RNA sequencing (scRNA-seq) and in vitro functional analyzes, we pinpoint Th2 cells, dendritic cells (DCs), and fibroblasts as crucial cell populations. The IL13-IL13RA1 ligand-receptor pair is identified as the most significant mediator of immune-stromal crosstalk in BP. Notably, fibroblasts and DCs expressing IL13RA1 respond to IL13-secreting Th2 cells, thereby amplifying Th2 cell-mediated cascade responses, which occurs through the specific upregulation of PLA2G2A in fibroblasts and CCL17 in myeloid cells, creating a positive feedback loop integral to immune-stromal crosstalk. Furthermore, PLA2G2A and CCL17 contribute to an increased titer of pathogenic anti-BP180-NC16A autoantibodies in BP patients. Our work provides a comprehensive insight into BP pathogenesis and shows a mechanism governing immune-stromal interactions, providing potential avenues for future therapeutic research.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interest.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

