Zeb1 mediates EMT/plasticity-associated ferroptosis sensitivity in cancer cells by regulating lipogenic enzyme expression and phospholipid composition
- PMID: 39009641
- PMCID: PMC11392809
- DOI: 10.1038/s41556-024-01464-1
Zeb1 mediates EMT/plasticity-associated ferroptosis sensitivity in cancer cells by regulating lipogenic enzyme expression and phospholipid composition
Abstract
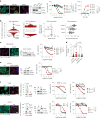
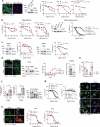
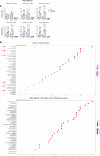
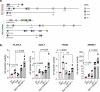
Therapy resistance and metastasis, the most fatal steps in cancer, are often triggered by a (partial) activation of the epithelial-mesenchymal transition (EMT) programme. A mesenchymal phenotype predisposes to ferroptosis, a cell death pathway exerted by an iron and oxygen-radical-mediated peroxidation of phospholipids containing polyunsaturated fatty acids. We here show that various forms of EMT activation, including TGFβ stimulation and acquired therapy resistance, increase ferroptosis susceptibility in cancer cells, which depends on the EMT transcription factor Zeb1. We demonstrate that Zeb1 increases the ratio of phospholipids containing pro-ferroptotic polyunsaturated fatty acids over cyto-protective monounsaturated fatty acids by modulating the differential expression of the underlying crucial enzymes stearoyl-Co-A desaturase 1 (SCD), fatty acid synthase (FASN), fatty acid desaturase 2 (FADS2), elongation of very long-chain fatty acid 5 (ELOVL5) and long-chain acyl-CoA synthetase 4 (ACSL4). Pharmacological inhibition of selected lipogenic enzymes (SCD and FADS2) allows the manipulation of ferroptosis sensitivity preferentially in high-Zeb1-expressing cancer cells. Our data are of potential translational relevance and suggest a combination of ferroptosis activators and SCD inhibitors for the treatment of aggressive cancers expressing high Zeb1.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

