Facile metabolic reprogramming distinguishes mycobacterial adaptation to hypoxia and starvation: ketosis drives starvation-induced persistence in M. bovis BCG
- PMID: 39009734
- PMCID: PMC11250799
- DOI: 10.1038/s42003-024-06562-2
Facile metabolic reprogramming distinguishes mycobacterial adaptation to hypoxia and starvation: ketosis drives starvation-induced persistence in M. bovis BCG
Abstract
Mycobacteria adapt to infection stresses by entering a reversible non-replicating persistence (NRP) with slow or no cell growth and broad antimicrobial tolerance. Hypoxia and nutrient deprivation are two well-studied stresses commonly used to model the NRP, yet little is known about the molecular differences in mycobacterial adaptation to these distinct stresses that lead to a comparable NRP phenotype. Here we performed a multisystem interrogation of the Mycobacterium bovis BCG (BCG) starvation response, which revealed a coordinated metabolic shift away from the glycolysis of nutrient-replete growth to depletion of lipid stores, lipolysis, and fatty acid ß-oxidation in NRP. This contrasts with BCG's NRP hypoxia response involving a shift to cholesterol metabolism and triglyceride storage. Our analysis reveals cryptic metabolic vulnerabilities of the starvation-induced NRP state, such as their newfound hypersensitivity to H2O2. These observations pave the way for developing precision therapeutics against these otherwise drug refractory pathogens.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
 ), S10 (
), S10 ( ), S20 (
), S20 ( ), and R6 (
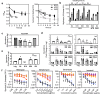
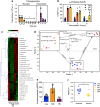
), and R6 ( ) BCG after 48 h of exposure. Data represent mean ± SD for n ≥ 6. c–e Data represent mean ± SD for n ≥ 3; *P < 0.05 determined by one-way ANOVA with Dunnett's test vs. Log. See Supplementary Data 4 for the source data used in these graphs.
) BCG after 48 h of exposure. Data represent mean ± SD for n ≥ 6. c–e Data represent mean ± SD for n ≥ 3; *P < 0.05 determined by one-way ANOVA with Dunnett's test vs. Log. See Supplementary Data 4 for the source data used in these graphs.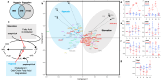


References
-
- Organization, W. H. Global Tuberculosis Report 2018. Report No. 978-92-4-156564-6, (World Health Organization, Geneva, 2018).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

