Reduced adipocyte glutaminase activity promotes energy expenditure and metabolic health
- PMID: 39009762
- PMCID: PMC11272588
- DOI: 10.1038/s42255-024-01083-y
Reduced adipocyte glutaminase activity promotes energy expenditure and metabolic health
Abstract
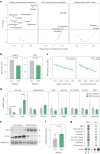
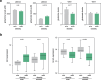
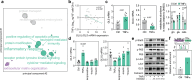
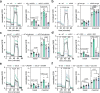
Glutamine and glutamate are interconverted by several enzymes and alterations in this metabolic cycle are linked to cardiometabolic traits. Herein, we show that obesity-associated insulin resistance is characterized by decreased plasma and white adipose tissue glutamine-to-glutamate ratios. We couple these stoichiometric changes to perturbed fat cell glutaminase and glutamine synthase messenger RNA and protein abundance, which together promote glutaminolysis. In human white adipocytes, reductions in glutaminase activity promote aerobic glycolysis and mitochondrial oxidative capacity via increases in hypoxia-inducible factor 1α abundance, lactate levels and p38 mitogen-activated protein kinase signalling. Systemic glutaminase inhibition in male and female mice, or genetically in adipocytes of male mice, triggers the activation of thermogenic gene programs in inguinal adipocytes. Consequently, the knockout mice display higher energy expenditure and improved glucose tolerance compared to control littermates, even under high-fat diet conditions. Altogether, our findings highlight white adipocyte glutamine turnover as an important determinant of energy expenditure and metabolic health.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











References
MeSH terms
Substances
Grants and funding
- 856404/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- 0064142/Novo Nordisk Fonden (Novo Nordisk Foundation)
- NNF20OC0061149/Novo Nordisk Fonden (Novo Nordisk Foundation)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

