Novel immunomodulatory properties of adenosine analogs promote their antiviral activity against SARS-CoV-2
- PMID: 39009832
- PMCID: PMC11315900
- DOI: 10.1038/s44319-024-00189-4
Novel immunomodulatory properties of adenosine analogs promote their antiviral activity against SARS-CoV-2
Abstract
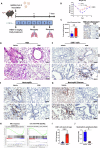
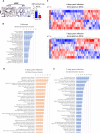
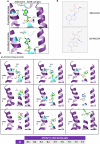
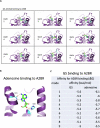
The COVID-19 pandemic reminded us of the urgent need for new antivirals to control emerging infectious diseases and potential future pandemics. Immunotherapy has revolutionized oncology and could complement the use of antivirals, but its application to infectious diseases remains largely unexplored. Nucleoside analogs are a class of agents widely used as antiviral and anti-neoplastic drugs. Their antiviral activity is generally based on interference with viral nucleic acid replication or transcription. Based on our previous work and computer modeling, we hypothesize that antiviral adenosine analogs, like remdesivir, have previously unrecognized immunomodulatory properties which contribute to their therapeutic activity. In the case of remdesivir, we here show that these properties are due to its metabolite, GS-441524, acting as an Adenosine A2A Receptor antagonist. Our findings support a new rationale for the design of next-generation antiviral agents with dual - immunomodulatory and intrinsic - antiviral properties. These compounds could represent game-changing therapies to control emerging viral diseases and future pandemics.
Keywords: Adenosine; Adenosine Analogs; Antivirals; Immunotherapy; Remdesivir.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
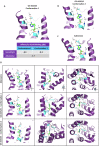
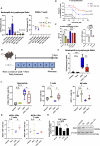
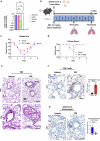
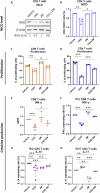


References
-
- Amstutz A, Speich B, Mentré F, Rueegg CS, Belhadi D, Assoumou L, Burdet C, Murthy S, Dodd LE, Wang Y et al (2023) Effects of remdesivir in patients hospitalised with COVID-19: a systematic review and individual patient data meta-analysis of randomised controlled trials. Lancet Respir Med 11:453–464 10.1016/S2213-2600(22)00528-8 - DOI - PMC - PubMed
-
- Barnes BJ, Adrover JM, Baxter-Stoltzfus A, Borczuk A, Cools-Lartigue J, Crawford JM, Daßler-Plenker J, Guerci P, Huynh C, Knight JS et al (2020) Targeting potential drivers of COVID-19: neutrophil extracellular trapsNeutrophil extracellular traps in COVID-19. J Exp Med 217:e20200652 10.1084/jem.20200652 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

