Unlocking opioid neuropeptide dynamics with genetically encoded biosensors
- PMID: 39009835
- PMCID: PMC11374718
- DOI: 10.1038/s41593-024-01697-1
Unlocking opioid neuropeptide dynamics with genetically encoded biosensors
Abstract
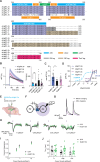
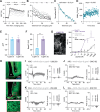
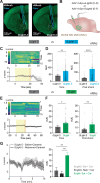
Neuropeptides are ubiquitous in the nervous system. Research into neuropeptides has been limited by a lack of experimental tools that allow for the precise dissection of their complex and diverse dynamics in a circuit-specific manner. Opioid peptides modulate pain, reward and aversion and as such have high clinical relevance. To illuminate the spatiotemporal dynamics of endogenous opioid signaling in the brain, we developed a class of genetically encoded fluorescence sensors based on kappa, delta and mu opioid receptors: κLight, δLight and µLight, respectively. We characterized the pharmacological profiles of these sensors in mammalian cells and in dissociated neurons. We used κLight to identify electrical stimulation parameters that trigger endogenous opioid release and the spatiotemporal scale of dynorphin volume transmission in brain slices. Using in vivo fiber photometry in mice, we demonstrated the utility of these sensors in detecting optogenetically driven opioid release and observed differential opioid release dynamics in response to fearful and rewarding conditions.
© 2024. The Author(s).
Conflict of interest statement
L.T. is a co-founder of Seven Biosciences. All other authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
- U19NS1237190/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R21EY031858/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R01 DA008863/DA/NIDA NIH HHS/United States
- R56 DA008863/DA/NIDA NIH HHS/United States
- R21 DA058300/DA/NIDA NIH HHS/United States
- R37 DA008863/DA/NIDA NIH HHS/United States
- U01 NS113295/NS/NINDS NIH HHS/United States
- ZIA MH002970/ImNIH/Intramural NIH HHS/United States
- R01 DK126740/DK/NIDDK NIH HHS/United States
- R37 DA033396/DA/NIDA NIH HHS/United States
- R61 DA051489/DA/NIDA NIH HHS/United States
- U01NS120820/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- U01 NS103522/NS/NINDS NIH HHS/United States
- U01 NS120820/NS/NINDS NIH HHS/United States

