This is a preprint.
Proteogenomics in cerebrospinal fluid and plasma reveals new biological fingerprint of cerebral small vessel disease
- PMID: 39011113
- PMCID: PMC11247936
- DOI: 10.21203/rs.3.rs-4535534/v1
Proteogenomics in cerebrospinal fluid and plasma reveals new biological fingerprint of cerebral small vessel disease
Update in
-
Proteogenomics in cerebrospinal fluid and plasma reveals new biological fingerprint of cerebral small vessel disease.Nat Aging. 2025 Dec;5(12):2514-2531. doi: 10.1038/s43587-025-01006-w. Epub 2025 Nov 20. Nat Aging. 2025. PMID: 41266628 Free PMC article.
Abstract
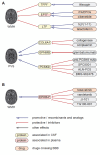
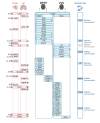
Cerebral small vessel disease (cSVD) is a leading cause of stroke and dementia with no specific mechanism-based treatment. We used Mendelian randomization to combine a unique cerebrospinal fluid (CSF) and plasma pQTL resource with the latest European-ancestry GWAS of MRI-markers of cSVD (white matter hyperintensities, perivascular spaces). We describe a new biological fingerprint of 49 protein-cSVD associations, predominantly in the CSF. We implemented a multipronged follow-up, across fluids, platforms, and ancestries (Europeans and East-Asian), including testing associations of direct plasma protein measurements with MRI-cSVD. We highlight 16 proteins robustly associated in both CSF and plasma, with 24/4 proteins identified in CSF/plasma only. cSVD-proteins were enriched in extracellular matrix and immune response pathways, and in genes enriched in microglia and specific microglial states (integration with single-nucleus RNA sequencing). Immune-related proteins were associated with MRI-cSVD already at age twenty. Half of cSVD-proteins were associated with stroke, dementia, or both, and seven cSVD-proteins are targets for known drugs (used for other indications in directions compatible with beneficial therapeutic effects. This first cSVD proteogenomic signature opens new avenues for biomarker and therapeutic developments.
Conflict of interest statement
Competing interests C.C. has received research support from GSK and EISAI and is a member of the advisory board of Circular Genomics and owns stocks in this company. CC is part of the scientific advisory board for ADmit. B.P. serves on the Steering Committee of the Yale Open Data Project funded by Johnson & Johnson. P.M.M. has received an honourarium as Chair of the UKRI Medical Research Council Neuroscience and Mental Health Board until March 2024. He acknowledges consultancy fees from, Biogen, Sudo. Nimbus and GSK. He has received speakers’ honoraria from Sanofi and Redburn, and has received research or educational funds from Biogen, Merck, Bristol Meyers Squibb and Nimbus. J.W declares no commercial COI; various academic research grants and is CI for LACunar Intervention Trials. The authors declared no potential conflicts of interest with respect to research, authorship, and/or publication of this article. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Figures




References
Publication types
Grants and funding
- P30 AG066444/AG/NIA NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- N01 HC085082/HL/NHLBI NIH HHS/United States
- P01 AG003991/AG/NIA NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- RF1 AG074007/AG/NIA NIH HHS/United States
- N01 HC055222/HL/NHLBI NIH HHS/United States
- N01 HC085079/HL/NHLBI NIH HHS/United States
- R00 AG062723/AG/NIA NIH HHS/United States
- 75N92021D00006/HL/NHLBI NIH HHS/United States
- R01 AG023629/AG/NIA NIH HHS/United States
- N01 HC085081/HL/NHLBI NIH HHS/United States
- R01 HL103612/HL/NHLBI NIH HHS/United States
- N01 HC085080/HL/NHLBI NIH HHS/United States
- U01 AG058922/AG/NIA NIH HHS/United States
- R01 AG044546/AG/NIA NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- P30 AG066515/AG/NIA NIH HHS/United States
- RF1 AG053303/AG/NIA NIH HHS/United States
- U01 HL130114/HL/NHLBI NIH HHS/United States
- HHSN268200800007C/HL/NHLBI NIH HHS/United States
- N01 HC085086/HL/NHLBI NIH HHS/United States
- N01 HC085083/HL/NHLBI NIH HHS/United States
- P01 AG026276/AG/NIA NIH HHS/United States
- RF1 AG058501/AG/NIA NIH HHS/United States
- R01 HL087652/HL/NHLBI NIH HHS/United States
- HHSN268201200036C/HL/NHLBI NIH HHS/United States
- HHSN268201800001C/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources

