Optimal linear ensemble of binary classifiers
- PMID: 39011276
- PMCID: PMC11249386
- DOI: 10.1093/bioadv/vbae093
Optimal linear ensemble of binary classifiers
Abstract
Motivation: The integration of vast, complex biological data with computational models offers profound insights and predictive accuracy. Yet, such models face challenges: poor generalization and limited labeled data.
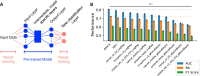
Results: To overcome these difficulties in binary classification tasks, we developed the Method for Optimal Classification by Aggregation (MOCA) algorithm, which addresses the problem of generalization by virtue of being an ensemble learning method and can be used in problems with limited or no labeled data. We developed both an unsupervised (uMOCA) and a supervised (sMOCA) variant of MOCA. For uMOCA, we show how to infer the MOCA weights in an unsupervised way, which are optimal under the assumption of class-conditioned independent classifier predictions. When it is possible to use labels, sMOCA uses empirically computed MOCA weights. We demonstrate the performance of uMOCA and sMOCA using simulated data as well as actual data previously used in Dialogue on Reverse Engineering and Methods (DREAM) challenges. We also propose an application of sMOCA for transfer learning where we use pre-trained computational models from a domain where labeled data are abundant and apply them to a different domain with less abundant labeled data.
Availability and implementation: GitHub repository, https://github.com/robert-vogel/moca.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
No competing interest is declared.
Figures



References
-
- Abadi M, Barham P, Chen J. et al. Tensorflow: a system for large-scale machine learning. In: 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI 16), Savannah, GA, USA, 2016, 265–83.
-
- Agarwal S, Graepel T, Herbrich R. et al. Generalization bounds for the area under the roc curve. J Mach Learn Res 2005;6:393–425.
-
- Ahsen ME, Vogel RM, Stolovitzky GA.. Unsupervised evaluation and weighted aggregation of ranked classification predictions. J Mach Learn Res 2019;20:1–40.
-
- Anders S, Huber W.. Differential Expression of RNA-seq Data at the Gene Level – The DESeq Package. Heidelberg, Germany: European Molecular Biology Laboratory (EMBL; ), 2012.
LinkOut - more resources
Full Text Sources

