Meta-transcriptomic analysis of companion animal infectomes reveals their diversity and potential roles in animal and human disease
- PMID: 39012105
- PMCID: PMC11351045
- DOI: 10.1128/msphere.00439-24
Meta-transcriptomic analysis of companion animal infectomes reveals their diversity and potential roles in animal and human disease
Abstract
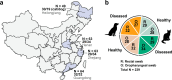
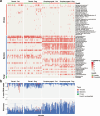
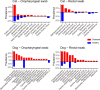
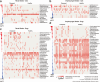
Companion animals such as cats and dogs harbor diverse microbial communities that can potentially impact human health due to close and frequent contact. To better characterize their total infectomes and assess zoonotic risks, we characterized the overall infectomes of companion animals (cats and dogs) and evaluated their potential zoonotic risks. Meta-transcriptomic analyses were performed on 239 samples from cats and dogs collected across China, identifying 24 viral species, 270 bacterial genera, and two fungal genera. Differences in the overall microbiome and infectome composition were compared across different animal species (cats or dogs), sampling sites (rectal or oropharyngeal), and health status (healthy or diseased). Diversity analyses revealed that viral abundance was generally higher in diseased animals compared to healthy ones, while differences in microbial composition were mainly driven by sampling site, followed by animal species and health status. Disease association analyses validated the pathogenicity of known pathogens and suggested potential pathogenic roles of previously undescribed bacteria and newly discovered viruses. Cross-species transmission analyses identified seven pathogens shared between cats and dogs, such as alphacoronavirus 1, which was detected in both oropharyngeal and rectal swabs albeit with differential pathogenicity. Further analyses showed that some viruses, like alphacoronavirus 1, harbored multiple lineages exhibiting distinct pathogenicity, tissue, or host preferences. Ultimately, a systematic evolutionary screening identified 27 potential zoonotic pathogens in this sample set, with far more bacterial than viral species, implying potential health threats to humans. Overall, our meta-transcriptomic analysis reveals a landscape of actively transcribing microorganisms in major companion animals, highlighting key pathogens, those with the potential for cross-species transmission, and possible zoonotic threats.
Importance: This study provides a comprehensive characterization of the entire community of infectious microbes (viruses, bacteria, and fungi) in companion animals like cats and dogs, termed the "infectome." By analyzing hundreds of samples from across China, the researchers identified numerous known and novel pathogens, including 27 potential zoonotic agents that could pose health risks to both animals and humans. Notably, some of these zoonotic pathogens were detected even in apparently healthy pets, highlighting the importance of surveillance. The study also revealed key microbial factors associated with respiratory and gastrointestinal diseases in pets, as well as potential cross-species transmission events between cats and dogs. Overall, this work sheds light on the complex microbial landscapes of companion animals and their potential impacts on animal and human health, underscoring the need for monitoring and management of these infectious agents.
Keywords: metagenomics; microbiome; veterinary medicine; virome; zoonoses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Demographic and zoological drivers of infectome diversity in companion cats with ascites.mSystems. 2024 Sep 17;9(9):e0063624. doi: 10.1128/msystems.00636-24. Epub 2024 Aug 9. mSystems. 2024. PMID: 39120143 Free PMC article.
-
[Pets (dogs/cats) as a possible source of opportunistic pathogenic fungi in humans].Klin Mikrobiol Infekc Lek. 2018 Jun;24(2):41-49. Klin Mikrobiol Infekc Lek. 2018. PMID: 30747431 Czech.
-
Disease Risk Assessments Involving Companion Animals: an Overview for 15 Selected Pathogens Taking a European Perspective.J Comp Pathol. 2016 Jul;155(1 Suppl 1):S75-97. doi: 10.1016/j.jcpa.2015.08.003. Epub 2015 Sep 28. J Comp Pathol. 2016. PMID: 26422413 Review.
-
Whole genome sequencing of Streptococcus suis revealed potential drug resistance and zoonotic transmission in companion cat.Trop Biomed. 2024 Mar 1;41(1):97-108. doi: 10.47665/tb.41.1.012. Trop Biomed. 2024. PMID: 38852139
-
Major Parasitic Zoonoses Associated with Dogs and Cats in Europe.J Comp Pathol. 2016 Jul;155(1 Suppl 1):S54-74. doi: 10.1016/j.jcpa.2015.10.179. Epub 2015 Dec 11. J Comp Pathol. 2016. PMID: 26687277 Review.
Cited by
-
In vitro modeling of feline gut fermentation: a comprehensive analysis of fecal microbiota and metabolic activity.Front Microbiol. 2025 Jan 29;16:1515865. doi: 10.3389/fmicb.2025.1515865. eCollection 2025. Front Microbiol. 2025. PMID: 39944651 Free PMC article.
-
Diverse viral pathogens in Australian canines: limited geographic structure and the first detection of an RNA virus in dingoes.Virus Evol. 2025 May 22;11(1):veaf042. doi: 10.1093/ve/veaf042. eCollection 2025. Virus Evol. 2025. PMID: 40584255 Free PMC article.
References
-
- Zhang N, Song C, Tao X, Zhu W. 2023. Epidemiologic features of human rabies in China from 2015-2021. Zoonoses 3. doi: 10.15212/ZOONOSES-2023-0012 - DOI
-
- Younus M, Wilkins MJ, Davies HD, Rahbar MH, Funk J, Nguyen C, Siddiqi AE, Cho S, Saeed AM. 2010. The role of exposures to animals and other risk factors in sporadic, non‐typhoidal Salmonella infections in Michigan children. Zoonoses Public Health 57:e170–e176. doi: 10.1111/j.1863-2378.2010.01324.x - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
