Comprehensive Assessment of Force-Field Performance in Molecular Dynamics Simulations of DNA/RNA Hybrid Duplexes
- PMID: 39012172
- PMCID: PMC11325551
- DOI: 10.1021/acs.jctc.4c00601
Comprehensive Assessment of Force-Field Performance in Molecular Dynamics Simulations of DNA/RNA Hybrid Duplexes
Abstract
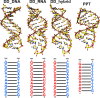
Mixed double helices formed by RNA and DNA strands, commonly referred to as hybrid duplexes or hybrids, are essential in biological processes like transcription and reverse transcription. They are also important for their applications in CRISPR gene editing and nanotechnology. Yet, despite their significance, the hybrid duplexes have been seldom modeled by atomistic molecular dynamics methodology, and there is no benchmark study systematically assessing the force-field performance. Here, we present an extensive benchmark study of polypurine tract (PPT) and Dickerson-Drew dodecamer hybrid duplexes using contemporary and commonly utilized pairwise additive and polarizable nucleic acid force fields. Our findings indicate that none of the available force-field choices accurately reproduces all the characteristic structural details of the hybrid duplexes. The AMBER force fields are unable to populate the C3'-endo (north) pucker of the DNA strand and underestimate inclination. The CHARMM force field accurately describes the C3'-endo pucker and inclination but shows base pair instability. The polarizable force fields struggle with accurately reproducing the helical parameters. Some force-field combinations even demonstrate a discernible conflict between the RNA and DNA parameters. In this work, we offer a candid assessment of the force-field performance for mixed DNA/RNA duplexes. We provide guidance on selecting utilizable force-field combinations and also highlight potential pitfalls and best practices for obtaining optimal performance.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Telesnitsky A.; Goff S., Reverse Transcriptase and the Generation of Retroviral DNA. In Retroviruses, Coffin J.; Hughes S.; Varmus H., Eds. Cold Spring Harbor Laboratory Press: New York, 1997. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
