Molecular Interaction Fields Describing Halogen Bond Formable Areas on Protein Surfaces
- PMID: 39012240
- PMCID: PMC11323840
- DOI: 10.1021/acs.jcim.4c00896
Molecular Interaction Fields Describing Halogen Bond Formable Areas on Protein Surfaces
Abstract
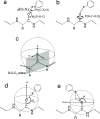
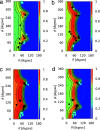
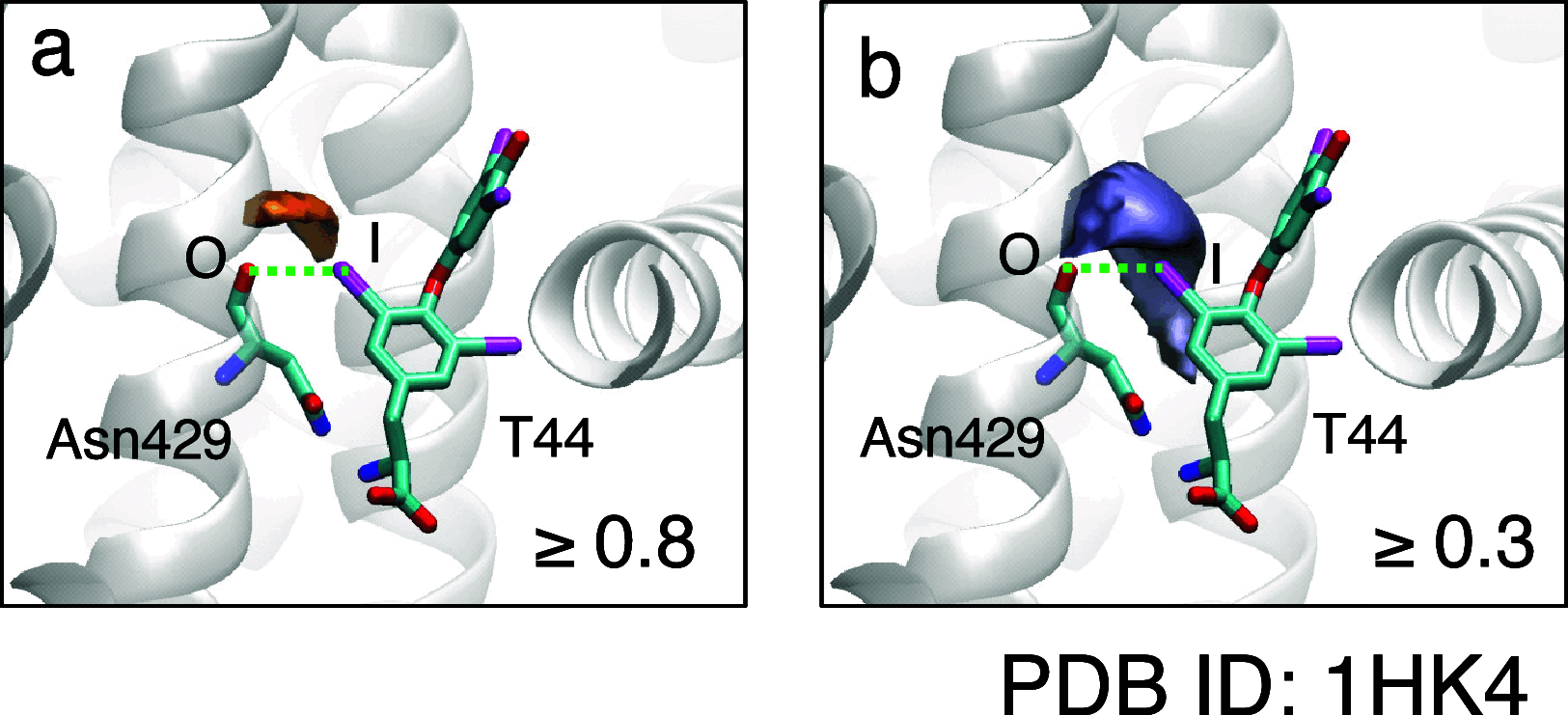
Molecular interaction fields (MIFs) are three-dimensional interaction maps that describe the intermolecular interactions expected to be formed around target molecules. In this paper, a method for the fast computation of MIFs using the approximation functions of quantum mechanics-level MIFs of small model molecules is proposed. MIF functions of N-methylacetamide with chlorobenzene, bromobenzene, and iodobenzene probes were precisely approximated and used to calculate the MIFs on protein surfaces. This method appropriately reproduced halogen-bond-formable areas around the ligand-binding sites of proteins, where halogen bond formation was suggested in a previous study.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

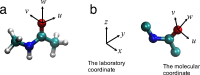

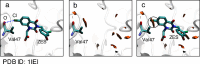

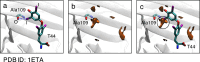
References
-
- Cruciani G.Molecular Interaction Fields; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, (2006). 10.1002/3527607676. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

