Development of an all-in-one real-time PCR assay for simultaneous detection of spotted fever group rickettsiae, severe fever with thrombocytopenia syndrome virus and hantaan virus prevalent in central China
- PMID: 39012922
- PMCID: PMC11280241
- DOI: 10.1371/journal.pntd.0012024
Development of an all-in-one real-time PCR assay for simultaneous detection of spotted fever group rickettsiae, severe fever with thrombocytopenia syndrome virus and hantaan virus prevalent in central China
Abstract
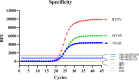
Central China has been reported to be one of the most important endemic areas of zoonotic infection by spotted fever group rickettsiae (SFGR), severe fever with thrombocytopenia syndrome virus (SFTSV) and hantaan virus (HTNV). Due to similar clinical symptoms, it is challenging to make a definite diagnosis rapidly and accurately in the absence of microbiological tests. In the present study, an all-in-one real-time PCR assay was developed for the simultaneous detection of nucleic acids from SFGR, SFTSV and HTNV. Three linear standard curves for determining SFGR-ompA, SFTSV-L and HTNV-L were obtained within the range of 101-106 copies/μL, with the PCR amplification efficiencies ranging from 93.46% to 96.88% and the regression coefficients R2 of >0.99. The detection limit was 1.108 copies/μL for SFGR-ompA, 1.075 copies/μL for SFTSV-L and 1.006 copies/μL for HTNV-L, respectively. Both the within-run and within-laboratory coefficients of variation on the cycle threshold (Ct) values were within the range of 0.53%-2.15%. It was also found there was no statistical difference in the Ct values between single template and multiple templates (PSFGR-ompA = 0.186, PSFTSV-L = 0.612, PHTNV-L = 0.298). The sensitivity, specificity, positive and negative predictive value were all 100% for determining SFGR-ompA and SFTSV-L, 97%, 100%, 100% and 99.6% for HTNV-L, respectively. Therefore, the all-in-one real-time PCR assay appears to be a reliable, sensitive, rapid, high-throughput and low cost-effective method to diagnose the zoonotic infection by SFGR, SFTSV and HTNV.
Copyright: © 2024 Wang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Ren YT, Tian HP, Xu JL, Liu MQ, Cai K, Chen SL, et al. Extensive genetic diversity of severe fever with thrombocytopenia syndrome virus circulating in Hubei Province, China, 2018–2022. PLoS Negl Trop Dis. 2023;17(9):e0011654. Epub 2023/09/18. doi: 10.1371/journal.pntd.0011654 ; PubMed Central PMCID: PMC10538666. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

