TLNRD1 is a CCM complex component and regulates endothelial barrier integrity
- PMID: 39013281
- PMCID: PMC11252447
- DOI: 10.1083/jcb.202310030
TLNRD1 is a CCM complex component and regulates endothelial barrier integrity
Abstract
We previously identified talin rod domain-containing protein 1 (TLNRD1) as a potent actin-bundling protein in vitro. Here, we report that TLNRD1 is expressed in the vasculature in vivo. Its depletion leads to vascular abnormalities in vivo and modulation of endothelial cell monolayer integrity in vitro. We demonstrate that TLNRD1 is a component of the cerebral cavernous malformations (CCM) complex through its direct interaction with CCM2, which is mediated by a hydrophobic C-terminal helix in CCM2 that attaches to a hydrophobic groove on the four-helix domain of TLNRD1. Disruption of this binding interface leads to CCM2 and TLNRD1 accumulation in the nucleus and actin fibers. Our findings indicate that CCM2 controls TLNRD1 localization to the cytoplasm and inhibits its actin-bundling activity and that the CCM2-TLNRD1 interaction impacts endothelial actin stress fiber and focal adhesion formation. Based on these results, we propose a new pathway by which the CCM complex modulates the actin cytoskeleton and vascular integrity.
© 2024 Ball et al.
Conflict of interest statement
Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. I. Paatero reported “University of Turku has registered trademark 3DFLUOHISTO, and I. Paatero is involved in the commercialization of 3DFLUOHISTO-technology.” No other disclosures were reported.
Figures

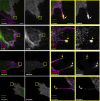
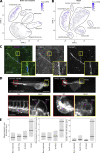
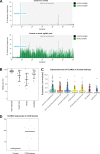

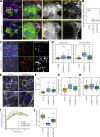




References
-
- Arzt, M., Deschamps J., Schmied C., Pietzsch T., Schmidt D., Tomancak P., Haase R., and Jug F.. 2022. LABKIT: Labeling and segmentation toolkit for big image data. Front. Comput. Sci. 4:777728. 10.3389/fcomp.2022.777728 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

