The WDR11 complex is a receptor for acidic-cluster-containing cargo proteins
- PMID: 39013469
- PMCID: PMC11316641
- DOI: 10.1016/j.cell.2024.06.024
The WDR11 complex is a receptor for acidic-cluster-containing cargo proteins
Abstract
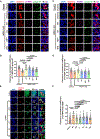
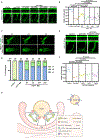
Vesicle trafficking is a fundamental process that allows for the sorting and transport of specific proteins (i.e., "cargoes") to different compartments of eukaryotic cells. Cargo recognition primarily occurs through coats and the associated proteins at the donor membrane. However, it remains unclear whether cargoes can also be selected at other stages of vesicle trafficking to further enhance the fidelity of the process. The WDR11-FAM91A1 complex functions downstream of the clathrin-associated AP-1 complex to facilitate protein transport from endosomes to the TGN. Here, we report the cryo-EM structure of human WDR11-FAM91A1 complex. WDR11 directly and specifically recognizes a subset of acidic clusters, which we term super acidic clusters (SACs). WDR11 complex assembly and its binding to SAC-containing proteins are indispensable for the trafficking of SAC-containing proteins and proper neuronal development in zebrafish. Our studies thus uncover that cargo proteins could be recognized in a sequence-specific manner downstream of a protein coat.
Keywords: AP-1; WDR11; cargo selection; neuronal development; vesicle tethering; vesicle trafficking.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

