Rational correction of pathogenic conformational defects in HTRA1
- PMID: 39013852
- PMCID: PMC11252331
- DOI: 10.1038/s41467-024-49982-8
Rational correction of pathogenic conformational defects in HTRA1
Abstract
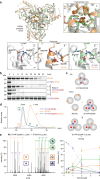
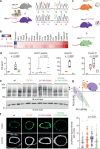
Loss-of-function mutations in the homotrimeric serine protease HTRA1 cause cerebral vasculopathy. Here, we establish independent approaches to achieve the functional correction of trimer assembly defects. Focusing on the prototypical R274Q mutation, we identify an HTRA1 variant that promotes trimer formation thus restoring enzymatic activity in vitro. Genetic experiments in Htra1R274Q mice further demonstrate that expression of this protein-based corrector in trans is sufficient to stabilize HtrA1-R274Q and restore the proteomic signature of the brain vasculature. An alternative approach employs supramolecular chemical ligands that shift the monomer-trimer equilibrium towards proteolytically active trimers. Moreover, we identify a peptidic ligand that activates HTRA1 monomers. Our findings open perspectives for tailored protein repair strategies.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests. Carsten Schmuck is deceased.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

