Rapid meiotic prophase chromosome movements in Arabidopsis thaliana are linked to essential reorganization at the nuclear envelope
- PMID: 39013853
- PMCID: PMC11252379
- DOI: 10.1038/s41467-024-50169-4
Rapid meiotic prophase chromosome movements in Arabidopsis thaliana are linked to essential reorganization at the nuclear envelope
Abstract
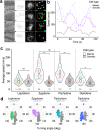
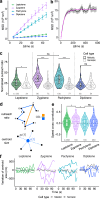
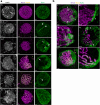
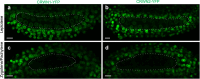
Meiotic rapid prophase chromosome movements (RPMs) require connections between the chromosomes and the cytoskeleton, involving SUN (Sad1/UNC-84)-domain-containing proteins at the inner nuclear envelope (NE). RPMs remain significantly understudied in plants, with respect to their importance in the regulation of meiosis. Here, we demonstrate that Arabidopsis thaliana meiotic centromeres undergo rapid (up to 500 nm/s) and uncoordinated movements during the zygotene and pachytene stages. These centromere movements are not affected by altered chromosome organization and recombination but are abolished in the double mutant sun1 sun2. We also document the changes in chromosome dynamics and nucleus organization during the transition from leptotene to zygotene, including telomere attachment to SUN-enriched NE domains, bouquet formation, and nucleolus displacement, all of which were defective in sun1 sun2. These results establish A. thaliana as a model species for studying the functional implications of meiotic RPMs and demonstrate the mechanistic conservation of telomere-led RPMs in plants.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

