Introducing carbon assimilation in yeasts using photosynthetic directed endosymbiosis
- PMID: 39013857
- PMCID: PMC11252298
- DOI: 10.1038/s41467-024-49585-3
Introducing carbon assimilation in yeasts using photosynthetic directed endosymbiosis
Abstract
Conversion of heterotrophic organisms into partially or completely autotrophic organisms is primarily accomplished by extensive metabolic engineering and laboratory evolution efforts that channel CO2 into central carbon metabolism. Here, we develop a directed endosymbiosis approach to introduce carbon assimilation in budding yeasts. Particularly, we engineer carbon assimilating and sugar-secreting photosynthetic cyanobacterial endosymbionts within the yeast cells, which results in the generation of yeast/cyanobacteria chimeras that propagate under photosynthetic conditions in the presence of CO2 and in the absence of feedstock carbon sources like glucose or glycerol. We demonstrate that the yeast/cyanobacteria chimera can be engineered to biosynthesize natural products under the photosynthetic conditions. Additionally, we expand our directed endosymbiosis approach to standard laboratory strains of yeasts, which transforms them into photosynthetic yeast/cyanobacteria chimeras. We anticipate that our studies will have significant implications for sustainable biotechnology, synthetic biology, and experimentally studying the evolutionary adaptation of an additional organelle in yeast.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

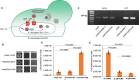
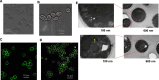
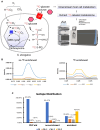
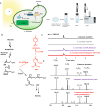
Comment in
-
Autotrophic yeast.Nat Commun. 2024 Jul 16;15(1):5948. doi: 10.1038/s41467-024-49586-2. Nat Commun. 2024. PMID: 39013898 Free PMC article.
References
-
- Claassens, N. J., Sousa, D. Z., dos Santos, V. A. P. M., de Vos, W. M. & van der Oost, J. Harnessing the power of microbial autotrophy. Nat. Rev. Microbiol.14, 692–706 (2016). - PubMed
-
- Zhuang, Z.-Y. & Li, S.-Y. Rubisco-based engineered Escherichia coli for in situ carbon dioxide recycling. Bioresour. Technol.150, 79–88 (2013). - PubMed
-
- Xia, P.-F. et al. Recycling carbon dioxide during xylose fermentation by engineered Saccharomyces cerevisiae. ACS Synth. Biol.6, 276–283 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

